You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000740_00414
You are here: Home > Sequence: MGYG000000740_00414
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp001917305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp001917305 | |||||||||||
| CAZyme ID | MGYG000000740_00414 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37345; End: 38445 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 48 | 321 | 8.2e-99 | 0.9963636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 1.58e-06 | 142 | 227 | 111 | 186 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 3.54e-06 | 76 | 287 | 37 | 236 | Cellulase (glycosyl hydrolase family 5). |
| COG3934 | COG3934 | 4.31e-04 | 35 | 187 | 2 | 148 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 7.69e-04 | 160 | 286 | 103 | 233 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANH83758.1 | 1.01e-110 | 17 | 362 | 42 | 390 |
| QDH79908.1 | 4.78e-108 | 3 | 362 | 7 | 388 |
| QNL50553.1 | 2.05e-107 | 17 | 359 | 40 | 386 |
| AHF14902.1 | 2.55e-107 | 16 | 362 | 37 | 386 |
| QUE52781.1 | 1.98e-106 | 13 | 362 | 17 | 373 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NIV9 | 4.47e-06 | 90 | 195 | 199 | 295 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
| Q2U2I3 | 4.47e-06 | 90 | 195 | 199 | 295 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
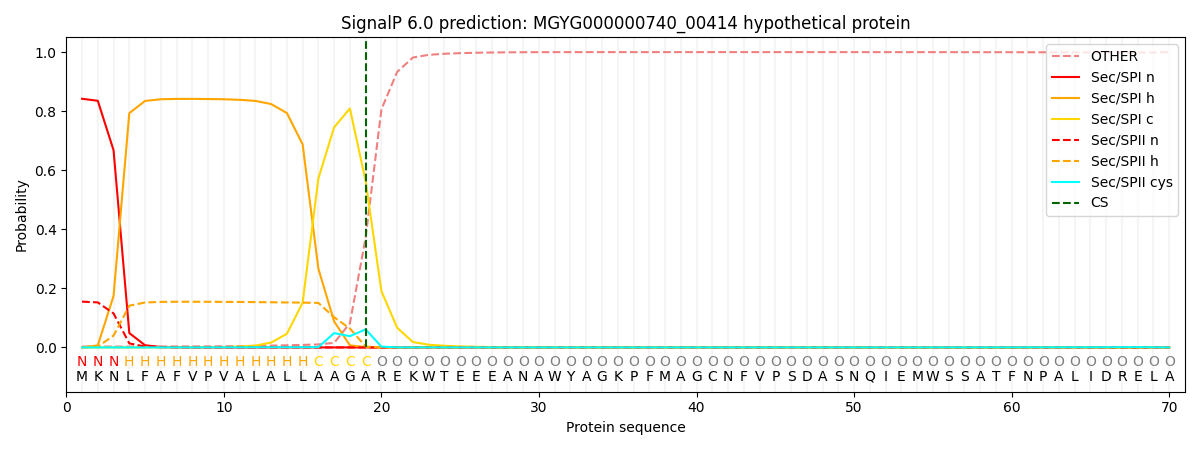
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006805 | 0.830825 | 0.161339 | 0.000350 | 0.000351 | 0.000318 |
