You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000740_01243
You are here: Home > Sequence: MGYG000000740_01243
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp001917305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp001917305 | |||||||||||
| CAZyme ID | MGYG000000740_01243 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33719; End: 35266 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 89 | 465 | 4.7e-45 | 0.966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.64e-34 | 171 | 465 | 17 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.28e-28 | 126 | 465 | 39 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 2.02e-26 | 173 | 465 | 62 | 308 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWI10666.1 | 3.25e-167 | 44 | 515 | 1 | 480 |
| AVM47074.1 | 3.27e-161 | 29 | 515 | 3 | 496 |
| QGA28189.1 | 1.33e-158 | 34 | 515 | 18 | 504 |
| QQZ02681.1 | 2.88e-158 | 44 | 514 | 25 | 494 |
| AHF92621.1 | 1.02e-150 | 42 | 505 | 16 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 3.10e-20 | 57 | 511 | 45 | 404 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 7.75e-19 | 57 | 511 | 45 | 404 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 5AY7_A | 9.00e-15 | 172 | 467 | 77 | 345 | Apsychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5AY7_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5D4Y_A A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples],5D4Y_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples] |
| 3EMC_A | 1.30e-14 | 171 | 465 | 62 | 327 | Crystalstructure of XynB, an intracellular xylanase from Paenibacillus barcinonensis [Paenibacillus barcinonensis],3EMQ_A Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor [Paenibacillus barcinonensis],3EMZ_A Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative [Paenibacillus barcinonensis] |
| 4L4O_A | 3.67e-13 | 171 | 465 | 69 | 334 | Thecrystal structure of CbXyn10B in native form [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY13 | 2.07e-14 | 167 | 472 | 86 | 380 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| O69231 | 7.21e-14 | 171 | 465 | 63 | 328 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| P23556 | 4.71e-12 | 160 | 465 | 63 | 339 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
| P49942 | 1.05e-10 | 260 | 460 | 144 | 363 | Endo-1,4-beta-xylanase A OS=Bacteroides ovatus OX=28116 GN=xylI PE=2 SV=1 |
| Q12603 | 1.22e-09 | 157 | 465 | 66 | 349 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
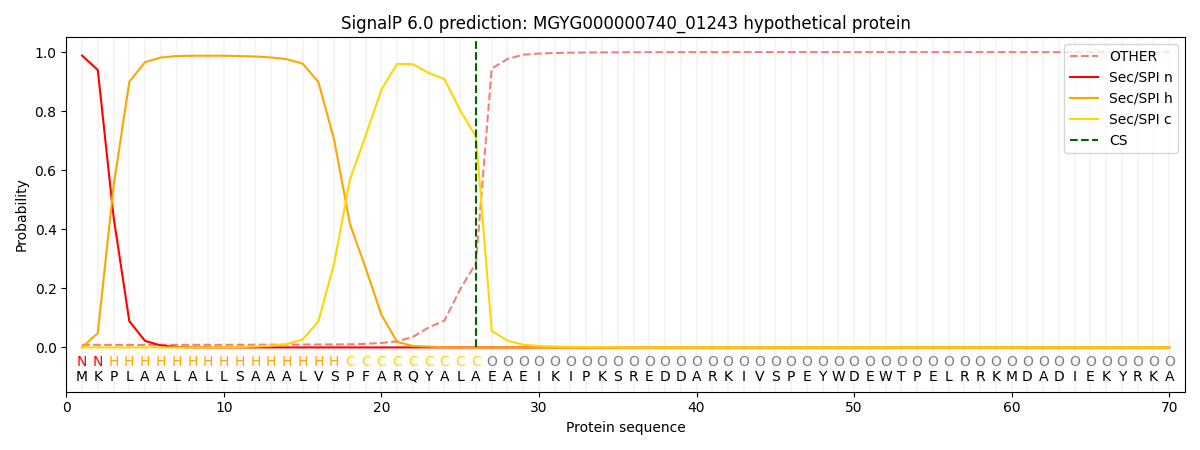
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.011988 | 0.983721 | 0.003396 | 0.000382 | 0.000257 | 0.000232 |
