You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000745_01031
You are here: Home > Sequence: MGYG000000745_01031
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
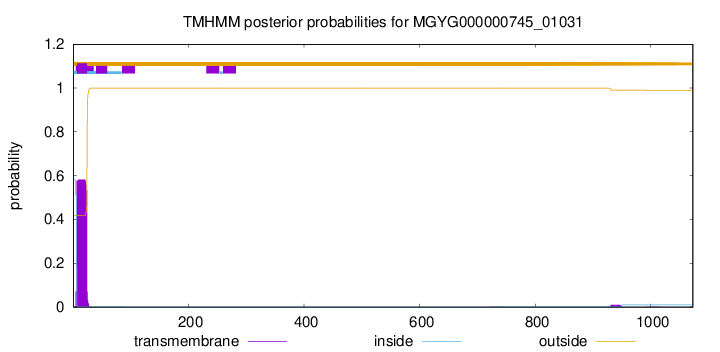
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; ; | |||||||||||
| CAZyme ID | MGYG000000745_01031 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8953; End: 12174 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 179 | 859 | 9.3e-82 | 0.699468085106383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.42e-39 | 179 | 554 | 39 | 402 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.02e-35 | 168 | 558 | 27 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.92e-21 | 211 | 550 | 111 | 454 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 6.55e-16 | 315 | 554 | 208 | 458 | beta-galactosidase. |
| pfam00703 | Glyco_hydro_2 | 1.43e-14 | 328 | 430 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK55911.1 | 4.20e-182 | 204 | 1066 | 383 | 1230 |
| QTH41860.1 | 1.17e-60 | 186 | 1031 | 23 | 858 |
| QOY90765.1 | 3.48e-48 | 175 | 1029 | 54 | 876 |
| QSZ05520.1 | 7.86e-48 | 209 | 949 | 55 | 782 |
| SDS96723.1 | 2.63e-47 | 181 | 1029 | 23 | 895 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 5.80e-26 | 187 | 599 | 67 | 475 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 3FN9_A | 7.22e-26 | 181 | 586 | 35 | 435 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 4JKM_A | 5.79e-24 | 177 | 704 | 39 | 589 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 6ECA_A | 6.13e-23 | 187 | 545 | 74 | 433 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 4JKK_A | 7.29e-23 | 187 | 544 | 43 | 404 | CrystalStructure of Streptococcus agalactiae beta-glucuronidase in space group I222 [Streptococcus agalactiae 2603V/R],4JKL_A Crystal Structure of Streptococcus agalactiae beta-glucuronidase in space group P21212 [Streptococcus agalactiae 2603V/R],4JKL_B Crystal Structure of Streptococcus agalactiae beta-glucuronidase in space group P21212 [Streptococcus agalactiae 2603V/R] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 2.18e-26 | 205 | 543 | 52 | 379 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| T2KPJ7 | 3.02e-19 | 180 | 558 | 77 | 438 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| Q4FAT7 | 8.08e-15 | 187 | 682 | 80 | 597 | Beta-glucuronidase OS=Sus scrofa OX=9823 GN=GUSB PE=3 SV=1 |
| P26257 | 1.17e-14 | 213 | 544 | 58 | 381 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| A5F5U6 | 1.50e-14 | 157 | 544 | 56 | 452 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
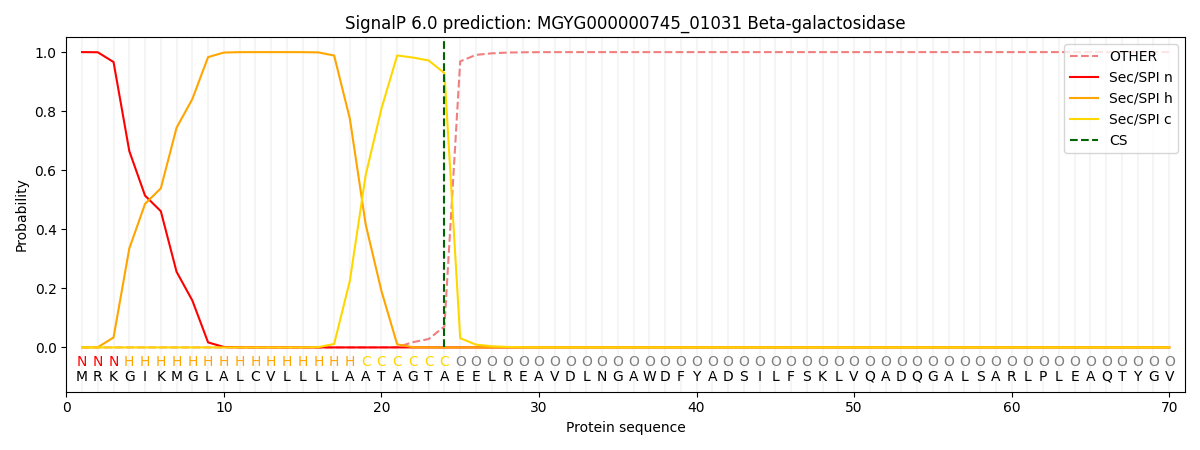
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000283 | 0.999031 | 0.000181 | 0.000177 | 0.000151 | 0.000146 |