You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000747_01665
You are here: Home > Sequence: MGYG000000747_01665
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; ; | |||||||||||
| CAZyme ID | MGYG000000747_01665 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2525; End: 4390 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 28 | 327 | 3.6e-30 | 0.8304093567251462 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd09083 | EEP-1 | 1.54e-77 | 370 | 617 | 1 | 251 | Exonuclease-Endonuclease-Phosphatase domain; uncharacterized family 1. This family of uncharacterized proteins belongs to a superfamily that includes the catalytic domain (exonuclease/endonuclease/phosphatase, EEP, domain) of a diverse set of proteins including the ExoIII family of apurinic/apyrimidinic (AP) endonucleases, inositol polyphosphate 5-phosphatases (INPP5), neutral sphingomyelinases (nSMases), deadenylases (such as the vertebrate circadian-clock regulated nocturnin), bacterial cytolethal distending toxin B (CdtB), deoxyribonuclease 1 (DNase1), the endonuclease domain of the non-LTR retrotransposon LINE-1, and related domains. These diverse enzymes share a common catalytic mechanism of cleaving phosphodiester bonds. Their substrates range from nucleic acids to phospholipids and perhaps, proteins. |
| cd15482 | Sialidase_non-viral | 2.71e-41 | 27 | 338 | 20 | 320 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 1.49e-25 | 33 | 334 | 1 | 276 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| cd08372 | EEP | 1.03e-16 | 373 | 617 | 3 | 240 | Exonuclease-Endonuclease-Phosphatase (EEP) domain superfamily. This large superfamily includes the catalytic domain (exonuclease/endonuclease/phosphatase or EEP domain) of a diverse set of proteins including the ExoIII family of apurinic/apyrimidinic (AP) endonucleases, inositol polyphosphate 5-phosphatases (INPP5), neutral sphingomyelinases (nSMases), deadenylases (such as the vertebrate circadian-clock regulated nocturnin), bacterial cytolethal distending toxin B (CdtB), deoxyribonuclease 1 (DNase1), the endonuclease domain of the non-LTR retrotransposon LINE-1, and related domains. These diverse enzymes share a common catalytic mechanism of cleaving phosphodiester bonds; their substrates range from nucleic acids to phospholipids and perhaps proteins. |
| COG3568 | ElsH | 2.19e-16 | 369 | 621 | 10 | 258 | Metal-dependent hydrolase, endonuclease/exonuclease/phosphatase family [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM08403.1 | 7.05e-106 | 3 | 339 | 9 | 344 |
| AOH43019.1 | 1.95e-78 | 20 | 337 | 21 | 334 |
| AEL26177.1 | 3.64e-72 | 7 | 347 | 28 | 352 |
| SDS49269.1 | 1.36e-71 | 11 | 354 | 24 | 360 |
| AVM46468.1 | 5.20e-67 | 2 | 364 | 3 | 359 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3G6S_A | 8.12e-27 | 368 | 617 | 3 | 257 | ChainA, Putative endonuclease/exonuclease/phosphatase family protein [Phocaeicola vulgatus ATCC 8482] |
| 3L1W_A | 3.88e-20 | 370 | 617 | 2 | 248 | Thecrystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis],3L1W_B The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis],3L1W_C The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis],3L1W_D The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis],3L1W_E The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis],3L1W_F The crystal structure of a functionally unknown conserved protein from Enterococcus faecalis V583 [Enterococcus faecalis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
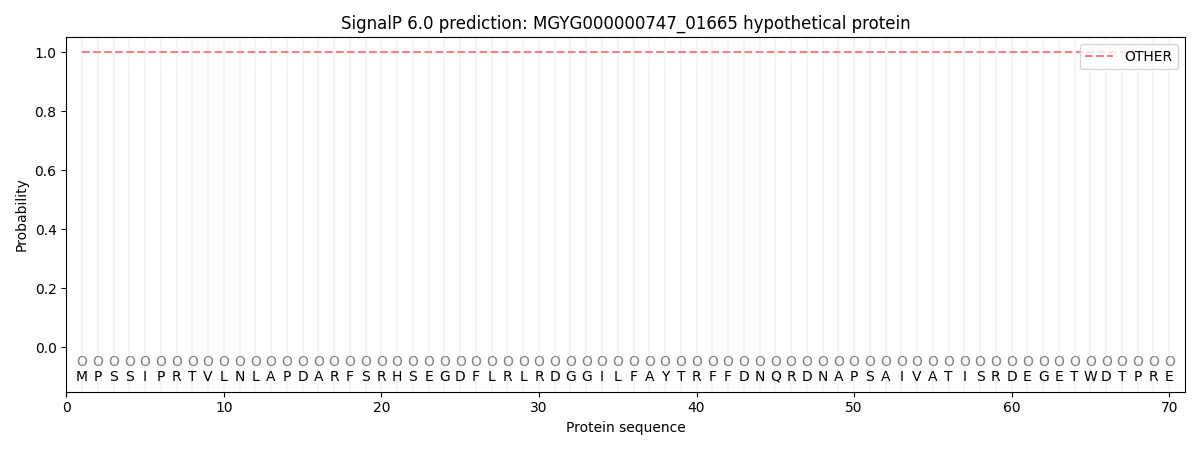
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000055 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
