You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000750_01210
You are here: Home > Sequence: MGYG000000750_01210
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; CAG-724; | |||||||||||
| CAZyme ID | MGYG000000750_01210 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7702; End: 9321 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 316 | 519 | 6e-29 | 0.8810572687224669 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 9.74e-38 | 296 | 538 | 157 | 386 | Predicted peptidase [General function prediction only]. |
| pfam13385 | Laminin_G_3 | 3.23e-15 | 149 | 291 | 16 | 149 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| smart00560 | LamGL | 2.26e-13 | 150 | 287 | 1 | 130 | LamG-like jellyroll fold domain. |
| COG3509 | LpqC | 3.91e-10 | 316 | 492 | 47 | 208 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| PRK10566 | PRK10566 | 6.15e-10 | 414 | 524 | 92 | 237 | esterase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61582.1 | 2.05e-45 | 316 | 537 | 825 | 1041 |
| QDU56037.1 | 3.65e-38 | 220 | 539 | 689 | 1007 |
| QJW99051.1 | 1.44e-33 | 306 | 539 | 34 | 240 |
| VTR91196.1 | 1.31e-32 | 285 | 539 | 14 | 239 |
| ABS60377.1 | 5.03e-29 | 300 | 539 | 9 | 245 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 1.95e-42 | 296 | 539 | 141 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 9.22e-19 | 316 | 539 | 21 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 4Q82_A | 6.84e-16 | 335 | 539 | 81 | 277 | CrystalStructure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365],4Q82_B Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365] |
| 4RGY_A | 2.65e-06 | 329 | 463 | 38 | 152 | Structuraland functional analysis of a low-temperature-active alkaline esterase from South China Sea marine sediment microbial metagenomic library [uncultured bacterium FLS12],4RGY_B Structural and functional analysis of a low-temperature-active alkaline esterase from South China Sea marine sediment microbial metagenomic library [uncultured bacterium FLS12] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
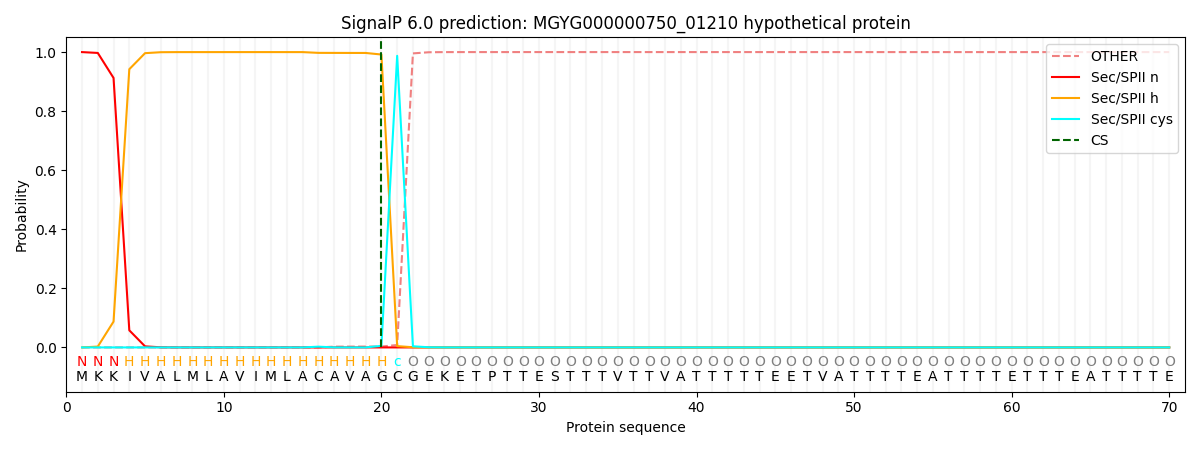
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000038 | 0.000000 | 0.000000 | 0.000000 |
