You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000759_00042
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium;
|
| CAZyme ID |
MGYG000000759_00042
|
| CAZy Family |
GT89 |
| CAZyme Description |
Terminal beta-(1->2)-arabinofuranosyltransferase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 564 |
|
62765.69 |
6.728 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000759 |
1836417 |
MAG |
Bangladesh |
Asia |
|
| Gene Location |
Start: 20050;
End: 21744
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GT89 |
16 |
563 |
4.3e-206 |
0.9784172661870504 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam11028
|
DUF2723 |
0.002 |
112 |
181 |
71 |
143 |
Protein of unknown function (DUF2723). This family is conserved in bacteria. The function is not known. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q8NLR0
|
1.58e-175 |
13 |
563 |
51 |
649 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=aftB PE=3 SV=1 |
|
O53582
|
1.22e-117 |
9 |
546 |
1 |
594 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=aftB PE=1 SV=3 |
This protein is predicted as OTHER
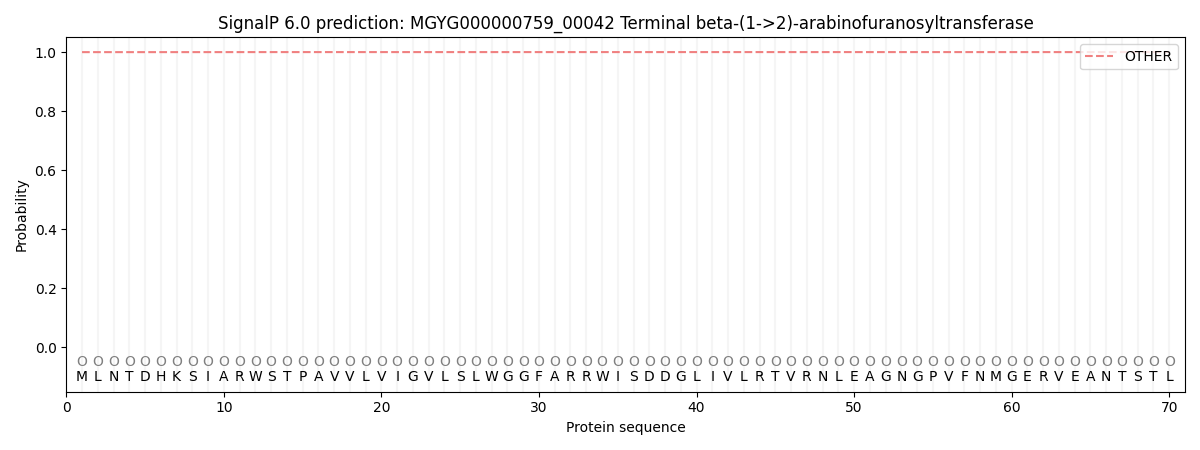
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999898
|
0.000080
|
0.000013
|
0.000001
|
0.000000
|
0.000046
|
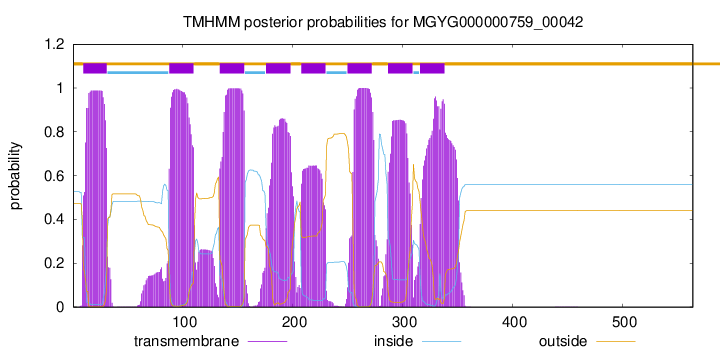
| start |
end |
| 10 |
31 |
| 88 |
110 |
| 134 |
156 |
| 176 |
198 |
| 208 |
230 |
| 250 |
272 |
| 287 |
309 |
| 316 |
338 |


