You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000767_01687
You are here: Home > Sequence: MGYG000000767_01687
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
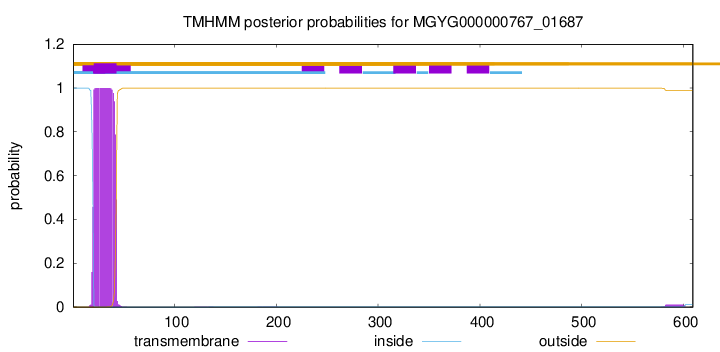
TMHMM annotations
Basic Information help
| Species | Paraeggerthella hongkongensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Eggerthellaceae; Paraeggerthella; Paraeggerthella hongkongensis | |||||||||||
| CAZyme ID | MGYG000000767_01687 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 2D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64; End: 1893 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 76 | 252 | 1.3e-62 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 2.07e-159 | 20 | 589 | 9 | 561 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 1.57e-124 | 18 | 589 | 4 | 681 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 9.65e-78 | 76 | 253 | 3 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| PRK11636 | mrcA | 1.06e-65 | 42 | 523 | 17 | 603 | penicillin-binding protein 1a; Provisional |
| COG4953 | PbpC | 2.66e-62 | 92 | 589 | 63 | 525 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACV56295.1 | 3.61e-258 | 1 | 589 | 1 | 603 |
| BAK44079.1 | 1.72e-256 | 1 | 589 | 1 | 588 |
| QOS67485.1 | 1.39e-254 | 1 | 589 | 1 | 608 |
| QTU84539.1 | 1.63e-247 | 1 | 589 | 1 | 590 |
| CBL03272.1 | 7.30e-224 | 6 | 607 | 1 | 599 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZG8_B | 6.22e-48 | 172 | 589 | 2 | 422 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 5FGZ_A | 4.54e-47 | 99 | 596 | 168 | 665 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 5.36e-47 | 99 | 596 | 189 | 686 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3DWK_A | 1.46e-43 | 66 | 589 | 7 | 547 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 3FWL_A | 2.42e-43 | 99 | 596 | 172 | 669 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7GHV1 | 1.78e-67 | 13 | 590 | 5 | 609 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 1.85e-67 | 13 | 590 | 5 | 609 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7FY32 | 1.85e-67 | 13 | 590 | 5 | 609 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| P70997 | 3.88e-65 | 69 | 553 | 57 | 531 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| Q68VU2 | 6.31e-65 | 99 | 596 | 79 | 686 | Penicillin-binding protein 1A OS=Rickettsia typhi (strain ATCC VR-144 / Wilmington) OX=257363 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
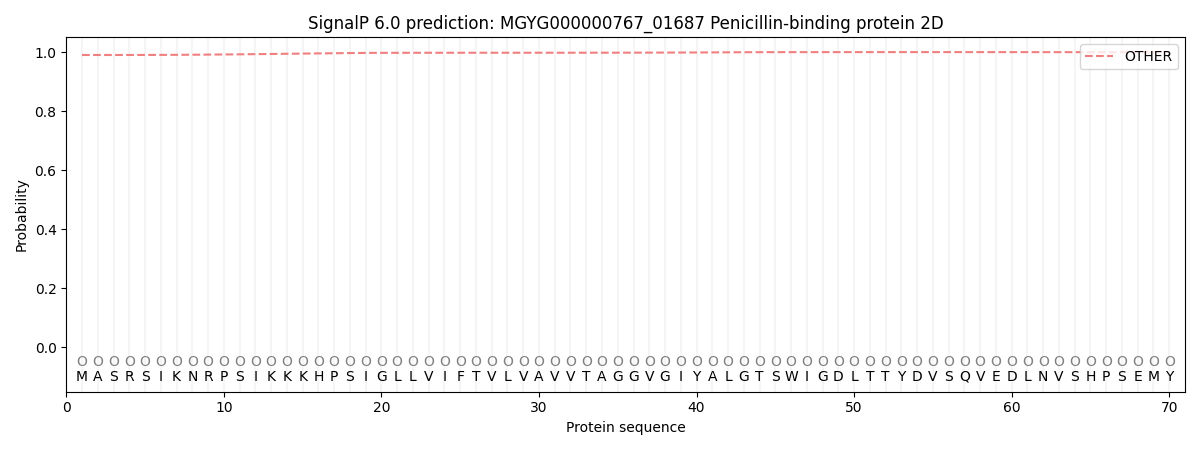
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.990726 | 0.008229 | 0.000076 | 0.000080 | 0.000032 | 0.000866 |