You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000771_01123
You are here: Home > Sequence: MGYG000000771_01123
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
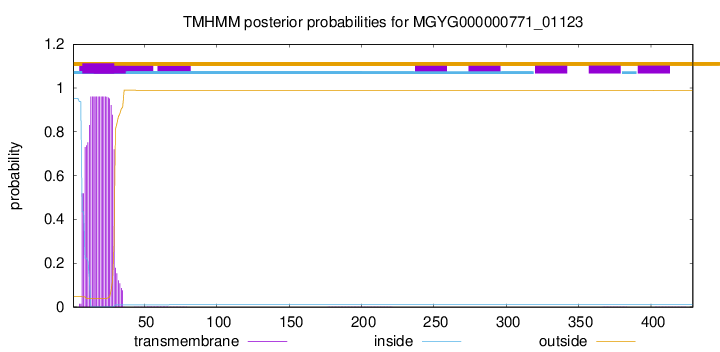
TMHMM annotations
Basic Information help
| Species | Lactococcus lactis_E | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus; Lactococcus lactis_E | |||||||||||
| CAZyme ID | MGYG000000771_01123 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1515; End: 2804 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 33 | 216 | 6.3e-38 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06415 | GH25_Cpl1-like | 5.49e-78 | 30 | 224 | 1 | 196 | Cpl-1 lysin (also known as Cpl-9 lysozyme / muramidase) is a bacterial cell wall endolysin encoded by the pneumococcal bacteriophage Cp-1, which cleaves the glycosidic N-acetylmuramoyl-(beta1,4)-N-acetylglucosamine bonds of the pneumococcal glycan chain, thus acting as an enzymatic antimicrobial agent (an enzybiotic) against streptococcal infections. Cpl-1 belongs to the CP family of lysozymes (CPL lysozymes) which includes the Cpl-7 lysin. Cpl-1 has a glycosyl hydrolase family 25 (GH25) catalytic domain with an irregular (beta/alpha)5-beta3 barrel and a C-terminal cell wall-anchoring module formed by six similar choline-binding repeats (ChBr's). The ChBr's facilitate the anchoring of Cpl-1 to the choline-containing teichoic acid of the pneumococcal cell wall. Other members of this domain family have an N-terminal CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain similar to that of the firmicute CHAP lysins and associated with endopeptidase activity. The Cpl-7 lysin is also included here as is LysB of Lactococcus phage, and the Mur lysin of Lactobacillus phage. |
| pfam01183 | Glyco_hydro_25 | 5.97e-34 | 33 | 217 | 1 | 180 | Glycosyl hydrolases family 25. |
| smart00641 | Glyco_25 | 8.42e-33 | 115 | 230 | 1 | 109 | Glycosyl hydrolases family 25. |
| cd00599 | GH25_muramidase | 4.83e-16 | 31 | 223 | 1 | 183 | Endo-N-acetylmuramidases (muramidases) are lysozymes (also referred to as peptidoglycan hydrolases) that degrade bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. This family of muramidases contains a glycosyl hydrolase family 25 (GH25) catalytic domain and is found in bacteria, fungi, slime molds, round worms, protozoans and bacteriophages. The bacteriophage members are referred to as endolysins which are involved in lysing the host cell at the end of the replication cycle to allow release of mature phage particles. Endolysins are typically modular enzymes consisting of a catalytically active domain that hydrolyzes the peptidoglycan cell wall and a cell wall-binding domain that anchors the protein to the cell wall. Endolysins generally have narrow substrate specificities with either intra-species or intra-genus bacteriolytic activity. |
| cd00118 | LysM | 8.74e-13 | 385 | 429 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTB96197.1 | 5.41e-311 | 1 | 429 | 1 | 429 |
| ARE18667.1 | 5.41e-311 | 1 | 429 | 1 | 429 |
| QTB94674.1 | 5.41e-311 | 1 | 429 | 1 | 429 |
| AXN65802.1 | 5.41e-311 | 1 | 429 | 1 | 429 |
| ARE26436.1 | 1.55e-310 | 1 | 429 | 1 | 429 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1OBA_A | 1.40e-23 | 67 | 210 | 37 | 178 | ChainA, Lysozyme [Streptococcus phage Cp1] |
| 1H09_A | 2.57e-23 | 67 | 210 | 36 | 177 | ChainA, LYSOZYME [Streptococcus phage Cp1] |
| 2IXU_A | 2.61e-23 | 67 | 210 | 37 | 178 | ChainA, LYSOZYME [Streptococcus phage Cp1] |
| 2IXV_A | 6.63e-23 | 67 | 210 | 37 | 178 | ChainA, LYSOZYME [Streptococcus phage Cp1],2J8F_A Chain A, LYSOZYME [Streptococcus phage Cp1],2J8G_A Chain A, LYSOZYME [Streptococcus phage Cp1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8HA43 | 1.18e-36 | 88 | 223 | 207 | 342 | D-alanyl-L-alanine endopeptidase OS=Streptococcus phage B30 OX=209152 PE=1 SV=2 |
| P19385 | 2.07e-25 | 75 | 210 | 46 | 178 | Lysozyme OS=Streptococcus phage Cp-7 OX=10748 GN=CPL7 PE=1 SV=2 |
| P19386 | 2.45e-24 | 67 | 210 | 37 | 178 | Lysozyme OS=Streptococcus phage Cp-9 OX=10749 GN=CPL9 PE=3 SV=1 |
| P15057 | 1.43e-22 | 67 | 210 | 37 | 178 | Lysozyme OS=Streptococcus phage Cp-1 OX=10747 GN=CPL1 PE=1 SV=2 |
| Q49UX4 | 1.43e-14 | 332 | 427 | 87 | 191 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
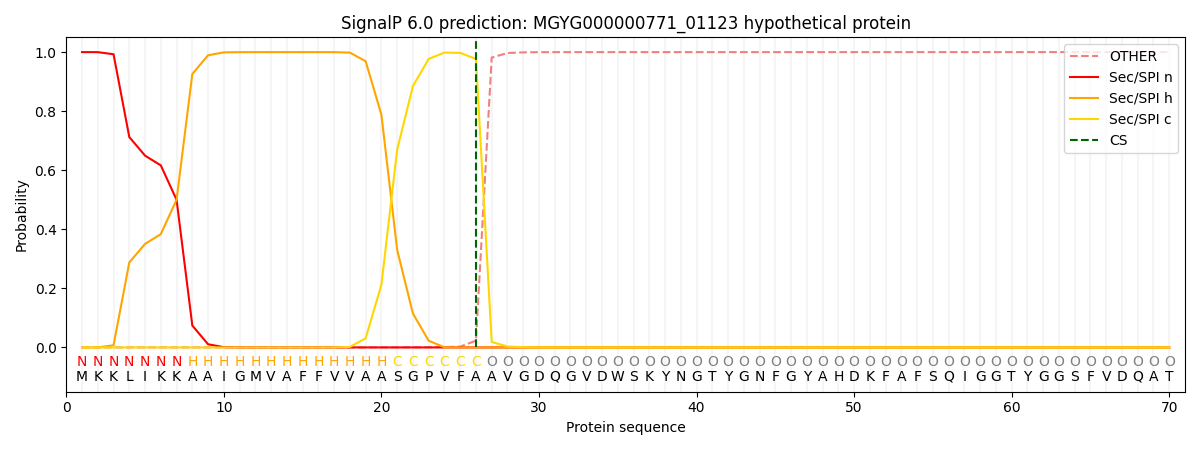
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.998881 | 0.000257 | 0.000184 | 0.000195 | 0.000174 |