You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000775_00574
You are here: Home > Sequence: MGYG000000775_00574
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
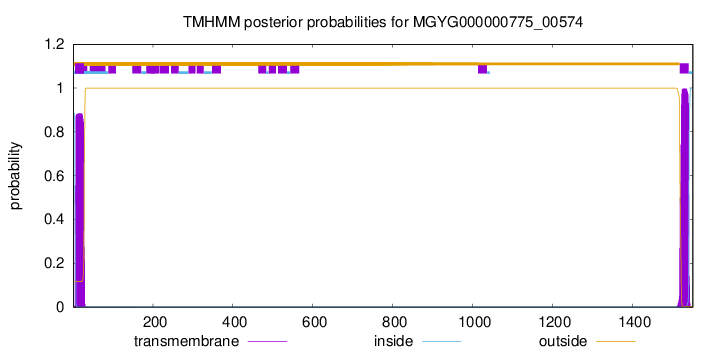
TMHMM annotations
Basic Information help
| Species | Bifidobacterium sp900551485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium sp900551485 | |||||||||||
| CAZyme ID | MGYG000000775_00574 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2303; End: 6961 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1021 | 1322 | 2.5e-98 | 0.9791666666666666 |
| GH28 | 153 | 533 | 3e-53 | 0.9476923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 4.69e-74 | 1021 | 1319 | 2 | 290 | Pectinesterase. |
| PLN02432 | PLN02432 | 5.52e-73 | 1021 | 1325 | 13 | 287 | putative pectinesterase |
| PLN02682 | PLN02682 | 2.10e-72 | 1003 | 1315 | 60 | 352 | pectinesterase family protein |
| PLN02773 | PLN02773 | 2.95e-70 | 1023 | 1321 | 9 | 293 | pectinesterase |
| COG5434 | Pgu1 | 5.18e-65 | 49 | 455 | 9 | 400 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATO41749.1 | 0.0 | 25 | 1439 | 30 | 1432 |
| AFU71934.1 | 0.0 | 25 | 1439 | 30 | 1432 |
| QYN60898.1 | 0.0 | 27 | 1439 | 35 | 1432 |
| QEY35289.1 | 0.0 | 38 | 1345 | 714 | 2020 |
| AYD48282.1 | 6.05e-84 | 41 | 535 | 31 | 486 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GQ8_A | 6.80e-46 | 1021 | 1324 | 9 | 301 | Pectinmethylesterase from Carrot [Daucus carota] |
| 2UVE_A | 3.01e-45 | 50 | 521 | 50 | 564 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 2NSP_A | 2.95e-41 | 1031 | 1325 | 18 | 335 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1XG2_A | 3.34e-40 | 1022 | 1306 | 6 | 277 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 3UW0_A | 2.60e-39 | 1022 | 1324 | 34 | 358 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SIJ9 | 1.11e-53 | 1023 | 1325 | 53 | 327 | Putative pectinesterase 11 OS=Arabidopsis thaliana OX=3702 GN=PME11 PE=3 SV=1 |
| Q9LVQ0 | 5.67e-52 | 1023 | 1318 | 9 | 290 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| P15922 | 1.82e-50 | 17 | 558 | 15 | 598 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| Q8VYZ3 | 2.85e-50 | 1029 | 1318 | 94 | 369 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
| Q42920 | 2.46e-49 | 1022 | 1318 | 133 | 417 | Pectinesterase/pectinesterase inhibitor OS=Medicago sativa OX=3879 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
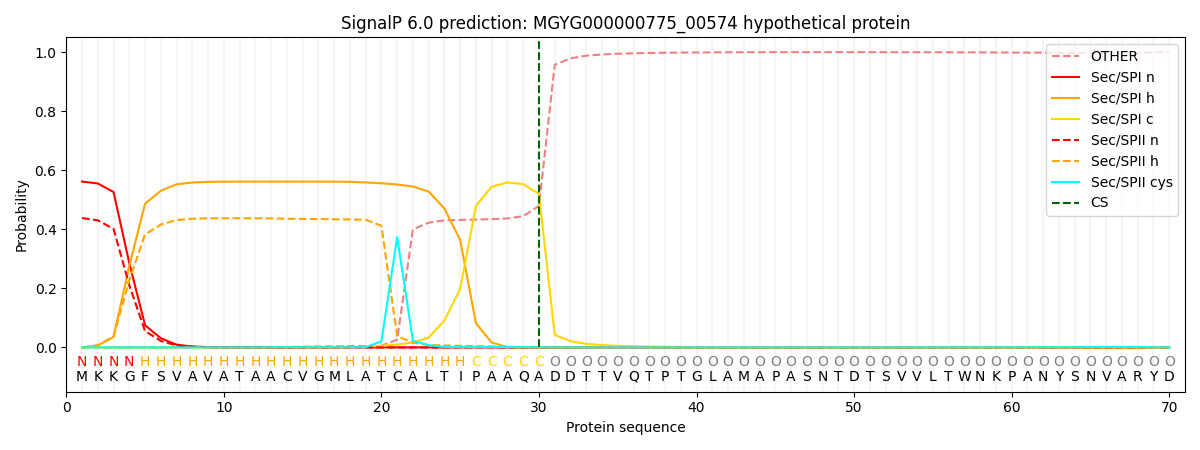
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000570 | 0.551874 | 0.446296 | 0.000754 | 0.000279 | 0.000202 |