You are browsing environment: HUMAN GUT
MGYG000000779_00394
Basic Information
help
Species
Prevotella sp000431975
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000431975
CAZyme ID
MGYG000000779_00394
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000779
2667664
MAG
Denmark
Europe
Gene Location
Start: 25672;
End: 27234
Strand: -
No EC number prediction in MGYG000000779_00394.
Family
Start
End
Evalue
family coverage
GT83
8
425
3.3e-66
0.7703703703703704
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
5.60e-22
8
430
15
435
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
COG2252
AzgA
0.006
299
424
324
436
Xanthine/uracil/vitamin C permease, AzgA family [Nucleotide transport and metabolism].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
2.21e-09
21
363
52
394
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
4.78e-17
10
381
5
367
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
A8FRR0
6.20e-08
5
369
12
368
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1
more
Q3KCC9
1.36e-06
14
322
37
337
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1
more
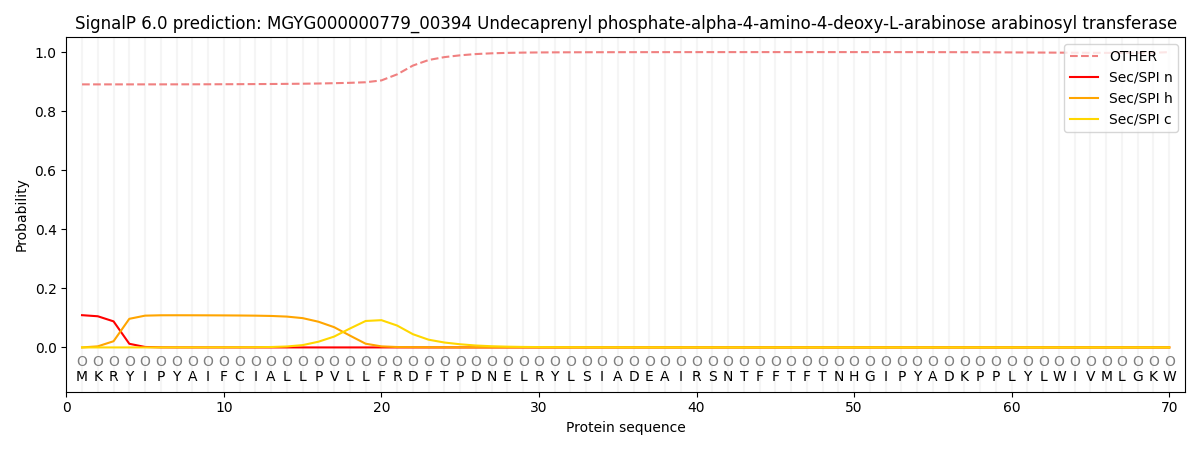
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.894162
0.104425
0.000620
0.000203
0.000188
0.000408
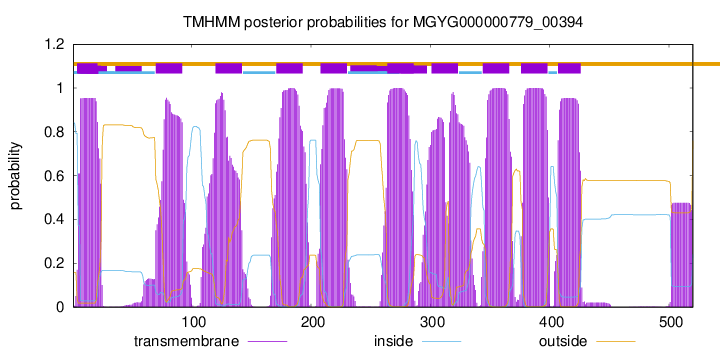
start
end
4
21
70
92
120
142
171
193
208
230
264
286
301
323
344
366
376
398
407
426