You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000782_01310
You are here: Home > Sequence: MGYG000000782_01310
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
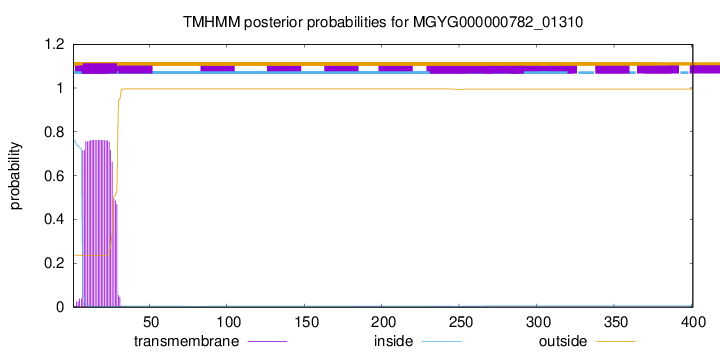
TMHMM annotations
Basic Information help
| Species | Phascolarctobacterium_A sp900541915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Phascolarctobacterium_A; Phascolarctobacterium_A sp900541915 | |||||||||||
| CAZyme ID | MGYG000000782_01310 | |||||||||||
| CAZy Family | GH171 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16360; End: 17565 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH171 | 47 | 400 | 2.9e-124 | 0.9971830985915493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07075 | DUF1343 | 2.10e-171 | 48 | 400 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| COG3876 | YbbC | 6.12e-142 | 9 | 400 | 8 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| cd16436 | beta_Kdo_transferase | 0.006 | 74 | 175 | 22 | 130 | beta-3-deoxy-D-manno-oct-2-ulosonic acid (Kdo)-transferase. KpsC and KpsS are retaining beta-3-deoxy-D-manno-oct-2-ulosonic acid (Kdo)-transferases. They are part of the ATP-binding cassette transporter dependent capsular polysaccharides (CPSs) synthesis pathway, one of two CPS synthesis pathways present in Escherichia coli. The poly-Kdo linker is thought to be the common feature of CPSs synthesized via this pathway. CPSs are high-molecular-mass cell-surface polysaccharides that are important virulence factors for many pathogenic bacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTV78142.1 | 4.47e-169 | 9 | 401 | 7 | 405 |
| QNP78041.1 | 1.12e-158 | 28 | 401 | 37 | 410 |
| BBG63384.1 | 1.59e-158 | 28 | 401 | 37 | 410 |
| AIF54236.1 | 4.71e-121 | 30 | 401 | 51 | 425 |
| AJQ29948.1 | 1.53e-118 | 30 | 400 | 48 | 421 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4K05_A | 7.35e-90 | 27 | 401 | 12 | 399 | Crystalstructure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343],4K05_B Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343] |
| 4JJA_A | 2.26e-50 | 28 | 276 | 6 | 257 | Crystalstructure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40407 | 7.37e-105 | 19 | 400 | 28 | 414 | Uncharacterized protein YbbC OS=Bacillus subtilis (strain 168) OX=224308 GN=ybbC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
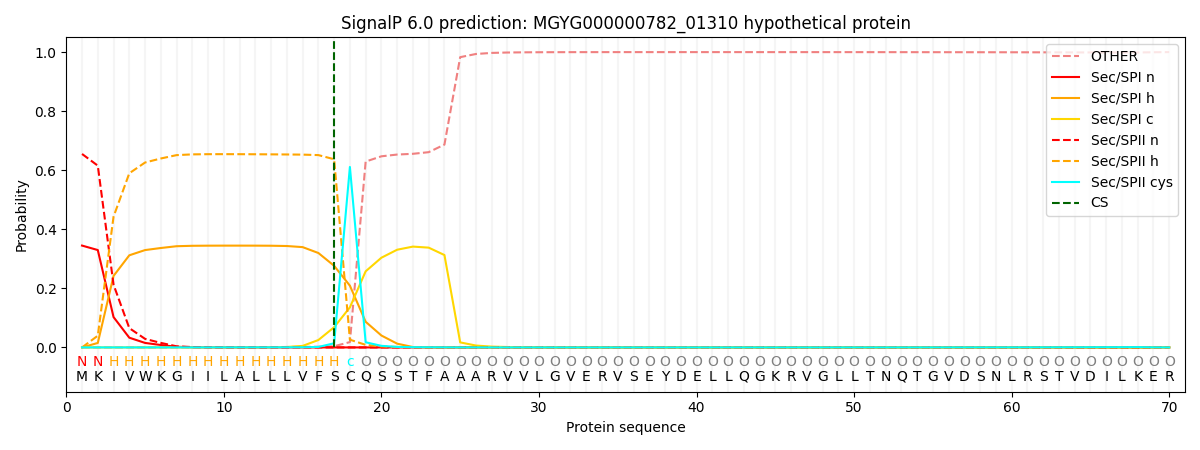
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000365 | 0.339074 | 0.660137 | 0.000141 | 0.000143 | 0.000127 |