You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000786_01598
You are here: Home > Sequence: MGYG000000786_01598
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Veillonella sp900545205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Veillonellaceae; Veillonella; Veillonella sp900545205 | |||||||||||
| CAZyme ID | MGYG000000786_01598 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1146; End: 2318 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 6 | 336 | 2.6e-48 | 0.6555555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 5.14e-20 | 2 | 215 | 158 | 376 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 2.29e-13 | 9 | 213 | 163 | 371 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBU37212.1 | 6.49e-279 | 2 | 389 | 155 | 542 |
| QQB17174.1 | 2.05e-266 | 2 | 390 | 155 | 543 |
| SNV02078.1 | 2.05e-266 | 2 | 390 | 155 | 543 |
| ACZ25367.1 | 2.05e-266 | 2 | 390 | 155 | 543 |
| CAB1277318.1 | 5.58e-264 | 2 | 390 | 155 | 543 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 7.31e-15 | 1 | 159 | 177 | 348 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4TBG8 | 1.61e-14 | 17 | 182 | 175 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella heidelberg (strain SL476) OX=454169 GN=arnT PE=3 SV=1 |
| B5R274 | 1.20e-11 | 17 | 182 | 175 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella enteritidis PT4 (strain P125109) OX=550537 GN=arnT PE=3 SV=1 |
| B5RCC6 | 1.20e-11 | 17 | 182 | 175 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella gallinarum (strain 287/91 / NCTC 13346) OX=550538 GN=arnT PE=3 SV=1 |
| B5FNU1 | 1.20e-11 | 17 | 182 | 175 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella dublin (strain CT_02021853) OX=439851 GN=arnT PE=3 SV=1 |
| B5EZI0 | 1.20e-11 | 17 | 182 | 175 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella agona (strain SL483) OX=454166 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000030 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
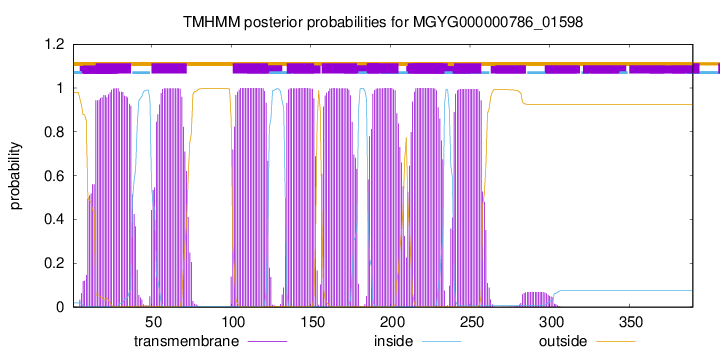
| start | end |
|---|---|
| 15 | 37 |
| 50 | 72 |
| 101 | 123 |
| 135 | 152 |
| 157 | 179 |
| 186 | 205 |
| 211 | 233 |
| 240 | 257 |
