You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000790_00067
You are here: Home > Sequence: MGYG000000790_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusobacterium nucleatum_E | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium nucleatum_E | |||||||||||
| CAZyme ID | MGYG000000790_00067 | |||||||||||
| CAZy Family | GH135 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16689; End: 18515 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH135 | 114 | 294 | 1.1e-23 | 0.7721518987341772 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12138 | Spherulin4 | 4.05e-21 | 110 | 295 | 41 | 235 | Spherulation-specific family 4. This protein is found in bacteria, archaea and eukaryotes. Proteins in this family are typically between 250 and 398 amino acids in length. There is a conserved NPG sequence motif and there are two completely conserved G residues that may be functionally important. Starvation will often induce spherulation - the production of spores - and this process may involve DNA-methylation. Changes in the methylation of spherulin4 are associated with the formation of spherules, but these changes are probably transient. Methylation of the gene accompanies its transcriptional activation, and spherulin4 mRNA is only detectable in late spherulating cultures and mature spherules. It is a spherulation-specific protein. |
| pfam03797 | Autotransporter | 3.59e-18 | 357 | 591 | 1 | 254 | Autotransporter beta-domain. Secretion of protein products occurs by a number of different pathways in bacteria. One of these pathways known as the type V pathway was first described for the IgA1 protease. The protein component that mediates secretion through the outer membrane is contained within the secreted protein itself, hence the proteins secreted in this way are called autotransporters. This family corresponds to the presumed integral membrane beta-barrel domain that transports the protein. This domain is found at the C-terminus of the proteins it occurs in. The N-terminus contains the variable passenger domain that is translocated across the membrane. Once the passenger domain is exported it is cleaved auto-catalytically in some proteins, in others a different protease is used and in some cases no cleavage occurs. |
| smart00869 | Autotransporter | 9.43e-16 | 359 | 598 | 1 | 267 | Autotransporter beta-domain. Secretion of protein products occurs by a number of different pathways in bacteria. One of these pathways known as the type IV pathway was first described for the IgA1 protease. The protein component that mediates secretion through the outer membrane is contained within the secreted protein itself, hence the proteins secreted in this way are called autotransporters. This family corresponds to the presumed integral membrane beta-barrel domain that transports the protein. This domain is found at the C-terminus of the proteins it occurs in. The N-terminus contains the variable passenger domain that is translocated across the membrane. Once the passenger domain is exported it is cleaved auto-catalytically in some proteins, in others a different peptidase is used and in some cases no cleavage occurs. |
| TIGR01414 | autotrans_barl | 2.04e-08 | 357 | 606 | 165 | 427 | outer membrane autotransporter barrel domain. A number of Gram-negative bacterial proteins, mostly found in pathogens and associated with virulence, contain a conserved C-terminal domain that integrates into the outer membrane and enables the N-terminal region to be delivered across the membrane. This C-terminal autotransporter domain is about 400 amino acids in length and includes the aromatic amino acid-rich OMP signal, typically ending with a Phe or Trp residue, at the extreme C-terminus. [Protein fate, Protein and peptide secretion and trafficking, Cellular processes, Pathogenesis] |
| pfam13505 | OMP_b-brl | 0.010 | 389 | 553 | 18 | 164 | Outer membrane protein beta-barrel domain. This domain is found in a wide range of outer membrane proteins. This domain assumes a membrane bound beta-barrel fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASQ47822.1 | 3.00e-241 | 34 | 608 | 142 | 707 |
| QUB96211.1 | 1.34e-236 | 34 | 608 | 139 | 704 |
| BBM45129.1 | 1.20e-157 | 56 | 603 | 219 | 760 |
| BBM57219.1 | 3.77e-156 | 56 | 603 | 219 | 760 |
| BBM53111.1 | 2.33e-67 | 1 | 431 | 1 | 451 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
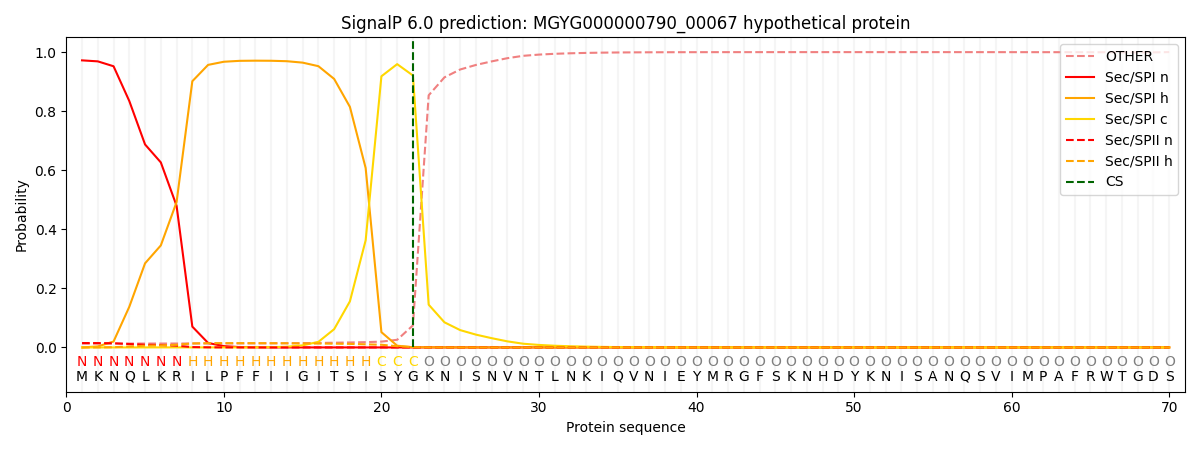
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.017057 | 0.965678 | 0.016271 | 0.000335 | 0.000310 | 0.000317 |
