You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000815_01339
You are here: Home > Sequence: MGYG000000815_01339
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas capnocytophagoides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas capnocytophagoides | |||||||||||
| CAZyme ID | MGYG000000815_01339 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 321547; End: 323733 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 452 | 724 | 2.5e-51 | 0.7656765676567657 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.60e-43 | 479 | 719 | 54 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 7.56e-41 | 474 | 724 | 91 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.98e-30 | 476 | 728 | 116 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 3.49e-09 | 55 | 142 | 5 | 92 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 7.69e-07 | 76 | 149 | 50 | 122 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA08088.1 | 7.65e-230 | 11 | 726 | 12 | 714 |
| AHW58788.1 | 3.95e-207 | 16 | 727 | 17 | 720 |
| APS38635.1 | 1.95e-204 | 13 | 728 | 14 | 709 |
| QEK50430.1 | 7.18e-189 | 1 | 726 | 1 | 711 |
| ADY53105.1 | 8.99e-189 | 1 | 727 | 1 | 719 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CPL_A | 1.20e-21 | 473 | 719 | 110 | 347 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 7CPK_A | 1.20e-21 | 473 | 719 | 110 | 347 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 6LPS_A | 4.69e-21 | 473 | 674 | 109 | 295 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2F8Q_A | 4.82e-21 | 473 | 642 | 107 | 263 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 2FGL_A | 4.88e-21 | 473 | 642 | 108 | 264 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2FGL_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07528 | 1.04e-20 | 473 | 719 | 154 | 391 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P40943 | 5.22e-19 | 477 | 700 | 154 | 352 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 PE=1 SV=1 |
| Q59675 | 6.47e-18 | 505 | 726 | 380 | 597 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
| P36917 | 3.10e-16 | 505 | 724 | 490 | 674 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P38535 | 3.97e-16 | 505 | 724 | 342 | 526 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
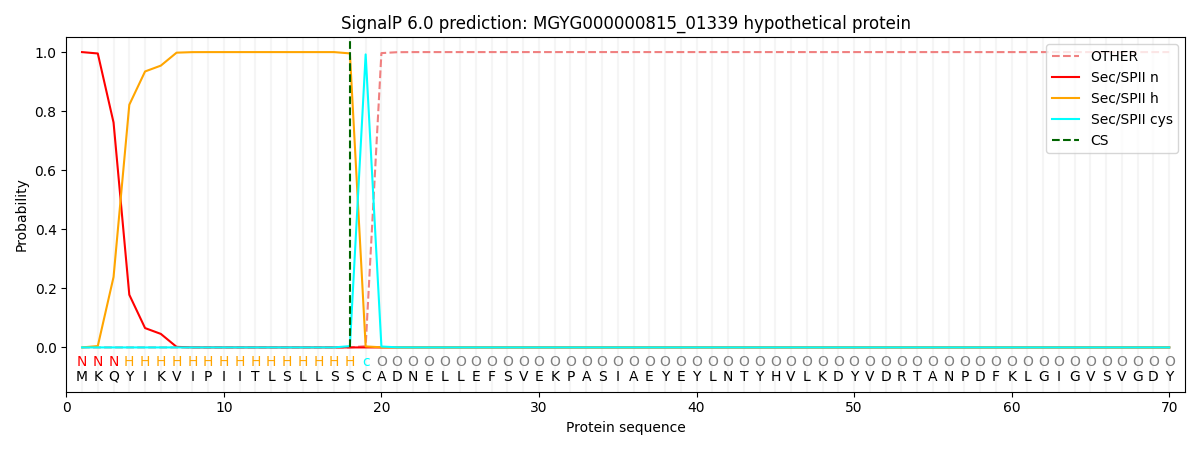
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000040 | 0.000000 | 0.000000 | 0.000000 |
