You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000815_01814
You are here: Home > Sequence: MGYG000000815_01814
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas capnocytophagoides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas capnocytophagoides | |||||||||||
| CAZyme ID | MGYG000000815_01814 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase 5A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 159073; End: 160035 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 44 | 276 | 7.8e-101 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.07e-58 | 41 | 284 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.03e-05 | 107 | 285 | 120 | 280 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK58758.1 | 4.48e-154 | 30 | 319 | 40 | 329 |
| AGL50932.1 | 1.82e-153 | 30 | 319 | 40 | 329 |
| QIK53341.1 | 3.66e-153 | 30 | 319 | 40 | 329 |
| QRQ63760.1 | 5.72e-135 | 26 | 317 | 33 | 324 |
| AWW31171.1 | 1.28e-134 | 33 | 316 | 36 | 319 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 1.14e-79 | 22 | 317 | 2 | 301 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZW_A | 2.00e-77 | 27 | 316 | 6 | 302 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 5IHS_A | 3.95e-77 | 26 | 317 | 28 | 321 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 1H11_A | 3.34e-76 | 24 | 317 | 3 | 301 | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYMEINTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION [Salipaludibacillus agaradhaerens],1H2J_A ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION [Salipaludibacillus agaradhaerens],1HF6_A ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE [Salipaludibacillus agaradhaerens],1OCQ_A COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine [Salipaludibacillus agaradhaerens],1W3K_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellobio Derived-tetrahydrooxazine [Salipaludibacillus agaradhaerens],1W3L_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellotri Derived-Tetrahydrooxazine [Salipaludibacillus agaradhaerens],4A3H_A 2',4' Dinitrophenyl-2-Deoxy-2-Fluro-B-D-Cellobioside Complex Of The Endoglucanase Cel5a From Bacillus Agaradhaerens At 1.6 A Resolution [Salipaludibacillus agaradhaerens],5A3H_A 2-Deoxy-2-Fluro-B-D-CellobiosylENZYME INTERMEDIATE COMPLEX Of The Endoglucanase Cel5a From Bacillus Agaradhearans At 1.8 Angstroms Resolution [Salipaludibacillus agaradhaerens],6A3H_A 2-Deoxy-2-Fluro-B-D-CellotriosylENZYME INTERMEDIATE COMPLEX OF THE Endoglucanase Cel5a From Bacillus Agaradhearans At 1.6 Angstrom Resolution [Salipaludibacillus agaradhaerens],7A3H_A Native Endoglucanase Cel5a Catalytic Core Domain At 0.95 Angstroms Resolution [Salipaludibacillus agaradhaerens],8A3H_A Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution [Salipaludibacillus agaradhaerens] |
| 1E5J_A | 3.55e-76 | 24 | 317 | 3 | 301 | EndoglucanaseCel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With Methyl-4ii-S-Alpha-Cellobiosyl-4ii-Thio Beta-Cellobioside [Salipaludibacillus agaradhaerens],1QHZ_A Native Tetragonal Structure Of The Endoglucanase Cel5a From Bacillus Agaradhaerens [Salipaludibacillus agaradhaerens],1QI0_A Endoglucanase Cel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With Cellobiose [Salipaludibacillus agaradhaerens],1QI2_A Endoglucanase Cel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With 2',4'-Dinitrophenyl 2-Deoxy-2-Fluoro-B- D-Cellotrioside [Salipaludibacillus agaradhaerens],2V38_A Family 5 endoglucanase Cel5A from Bacillus agaradhaerens in complex with cellobio-derived noeuromycin [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O85465 | 1.12e-74 | 10 | 317 | 6 | 327 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P06565 | 4.51e-73 | 26 | 317 | 31 | 327 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| P07983 | 4.62e-72 | 27 | 320 | 36 | 333 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 1.41e-70 | 27 | 316 | 36 | 329 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P15704 | 2.20e-70 | 31 | 316 | 45 | 335 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
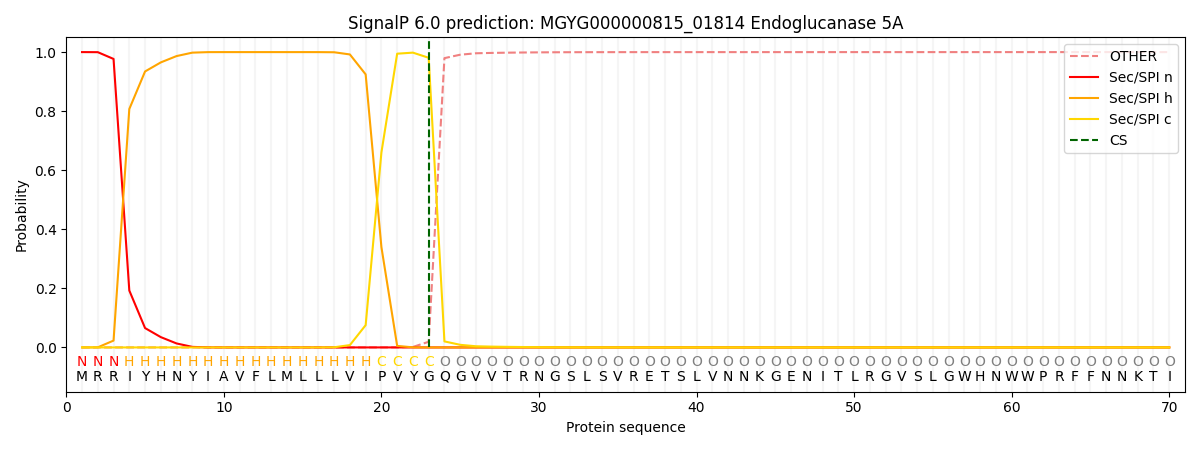
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000256 | 0.999132 | 0.000152 | 0.000156 | 0.000143 | 0.000133 |