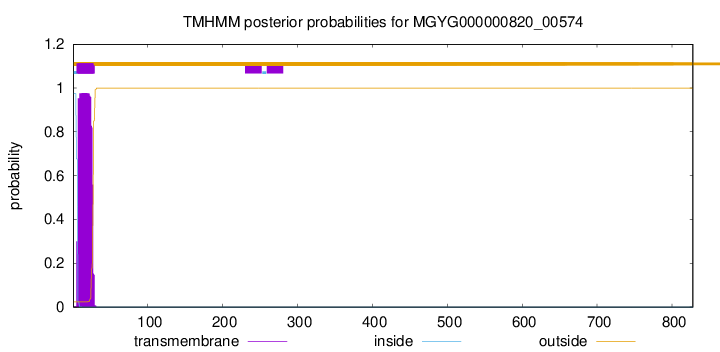
You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000820_00574
Basic Information
help
| Species |
UBA9506 sp003506415
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA9506; UBA9506; UBA9506 sp003506415
|
| CAZyme ID |
MGYG000000820_00574
|
| CAZy Family |
GH141 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000820 |
2712076 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 90131;
End: 92617
Strand: +
|
No EC number prediction in MGYG000000820_00574.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
153 |
657 |
1.8e-120 |
0.9829222011385199 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
4.05e-08 |
459 |
655 |
3 |
157 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
This protein is predicted as SP
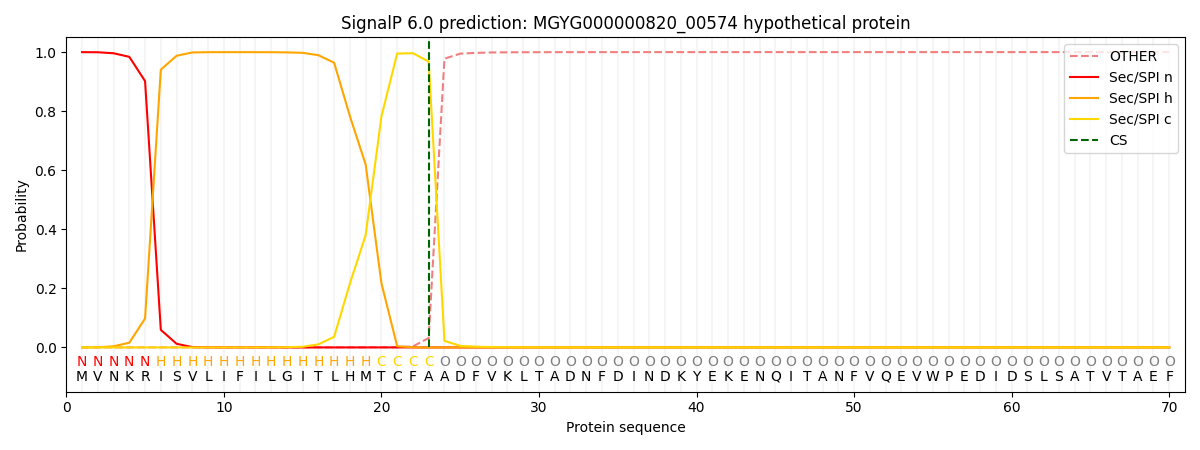
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.001501
|
0.997352
|
0.000367
|
0.000268
|
0.000246
|
0.000235
|