You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000820_02198
You are here: Home > Sequence: MGYG000000820_02198
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
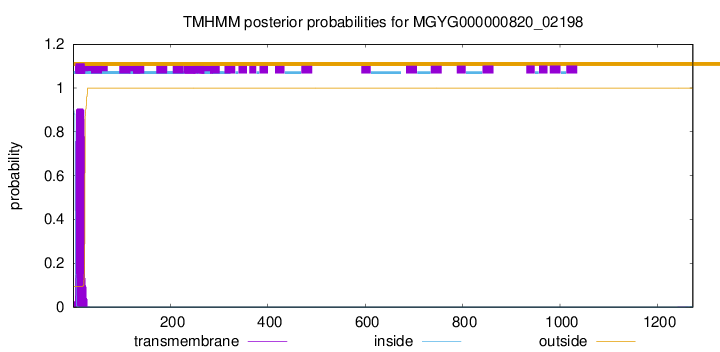
TMHMM annotations
Basic Information help
| Species | UBA9506 sp003506415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA9506; UBA9506; UBA9506 sp003506415 | |||||||||||
| CAZyme ID | MGYG000000820_02198 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47059; End: 50880 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 583 | 1101 | 3.9e-127 | 0.9867172675521821 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02018 | CBM_4_9 | 3.08e-09 | 211 | 348 | 1 | 128 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam13229 | Beta_helix | 7.26e-04 | 894 | 1041 | 1 | 112 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUL54275.1 | 4.45e-140 | 550 | 1180 | 366 | 979 |
| AIQ43746.1 | 5.99e-139 | 537 | 1270 | 348 | 1069 |
| QYR21751.1 | 3.77e-136 | 581 | 1181 | 408 | 992 |
| ANE49726.1 | 1.35e-116 | 579 | 1180 | 34 | 628 |
| SDS22682.1 | 1.62e-102 | 563 | 1181 | 19 | 607 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 1.05e-40 | 581 | 1115 | 27 | 605 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
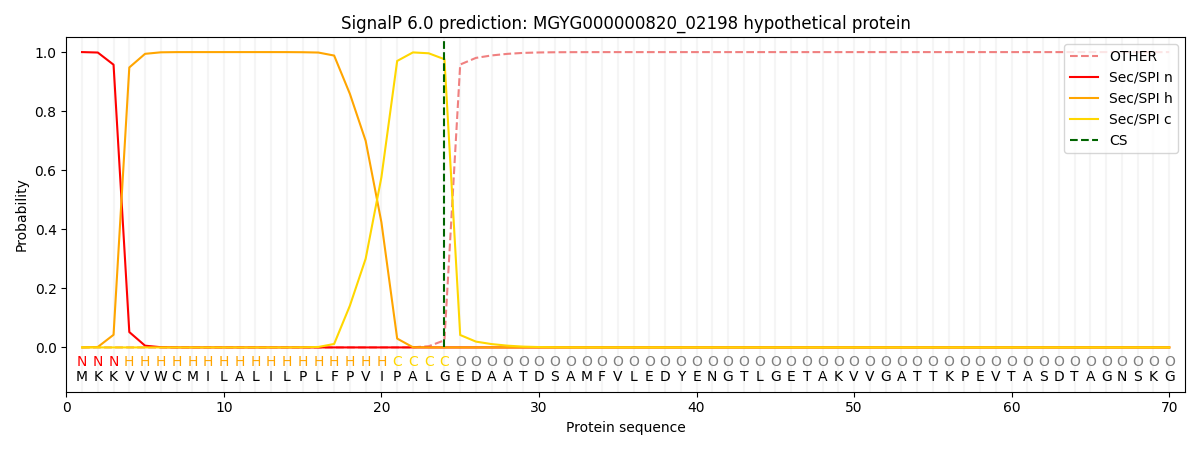
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000300 | 0.999088 | 0.000187 | 0.000155 | 0.000141 | 0.000141 |