You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000821_00025
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Sutterella;
|
| CAZyme ID |
MGYG000000821_00025
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000821 |
2141919 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 23656;
End: 25374
Strand: +
|
No EC number prediction in MGYG000000821_00025.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
18 |
491 |
3.7e-16 |
0.8148148148148148 |
MGYG000000821_00025 has no CDD domain.
MGYG000000821_00025 has no CAZyme hit.
This protein is predicted as OTHER
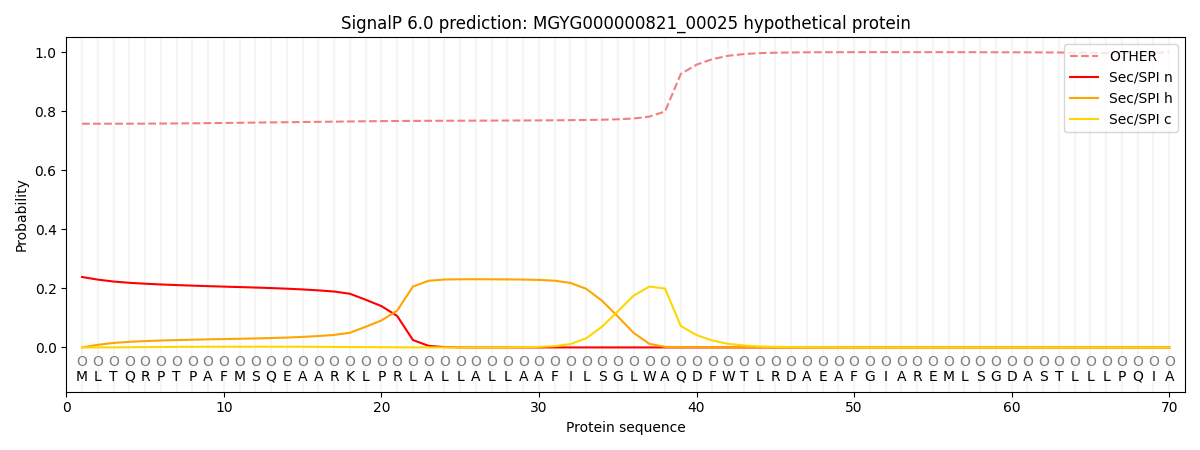
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.766987
|
0.225515
|
0.003364
|
0.000367
|
0.000433
|
0.003341
|
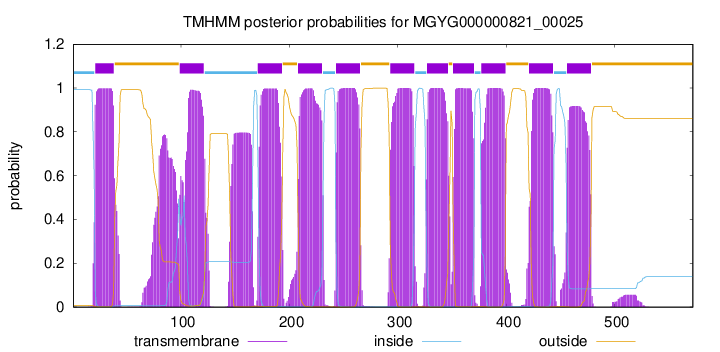
| start |
end |
| 21 |
38 |
| 99 |
121 |
| 171 |
193 |
| 208 |
230 |
| 243 |
265 |
| 293 |
315 |
| 327 |
346 |
| 351 |
370 |
| 377 |
399 |
| 421 |
443 |
| 456 |
478 |


