You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000823_00755
You are here: Home > Sequence: MGYG000000823_00755
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900550375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900550375 | |||||||||||
| CAZyme ID | MGYG000000823_00755 | |||||||||||
| CAZy Family | GH144 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21821; End: 22795 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH144 | 151 | 307 | 1.6e-63 | 0.3886138613861386 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5368 | COG5368 | 1.68e-27 | 150 | 301 | 6 | 163 | Uncharacterized protein [Function unknown]. |
| pfam13205 | Big_5 | 5.84e-10 | 55 | 131 | 18 | 95 | Bacterial Ig-like domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL13535.1 | 2.36e-182 | 1 | 303 | 1 | 302 |
| BBL04143.1 | 6.74e-182 | 1 | 303 | 1 | 302 |
| QUT88516.1 | 1.71e-96 | 10 | 303 | 14 | 300 |
| ALJ60472.1 | 1.09e-95 | 10 | 303 | 6 | 292 |
| SCD21374.1 | 3.90e-95 | 14 | 310 | 18 | 309 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4GL3_A | 7.92e-48 | 151 | 303 | 12 | 166 | Crystalstructure of a putative glucoamylase (BACUNI_03963) from Bacteroides uniformis ATCC 8492 at 2.01 A resolution [Bacteroides uniformis ATCC 8492] |
| 5GZH_A | 5.91e-41 | 152 | 303 | 24 | 182 | Endo-beta-1,2-glucanasefrom Chitinophaga pinensis - ligand free form [Chitinophaga pinensis DSM 2588],5GZH_B Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form [Chitinophaga pinensis DSM 2588] |
| 5GZK_A | 7.21e-41 | 152 | 303 | 44 | 202 | Endo-beta-1,2-glucanasefrom Chitinophaga pinensis - sophorotriose and glucose complex [Chitinophaga pinensis DSM 2588],5GZK_B Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex [Chitinophaga pinensis DSM 2588] |
| 5Z06_A | 6.80e-40 | 152 | 303 | 282 | 434 | Crystalstructure of beta-1,2-glucanase from Parabacteroides distasonis [Parabacteroides distasonis ATCC 8503],5Z06_B Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis [Parabacteroides distasonis ATCC 8503] |
| 4QT9_A | 1.38e-38 | 152 | 303 | 19 | 177 | Crystalstructure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185],4QT9_B Crystal structure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6LGF6 | 3.89e-39 | 152 | 303 | 299 | 451 | Exo beta-1,2-glucooligosaccharide sophorohydrolase (non-reducing end) OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=BDI_3064 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
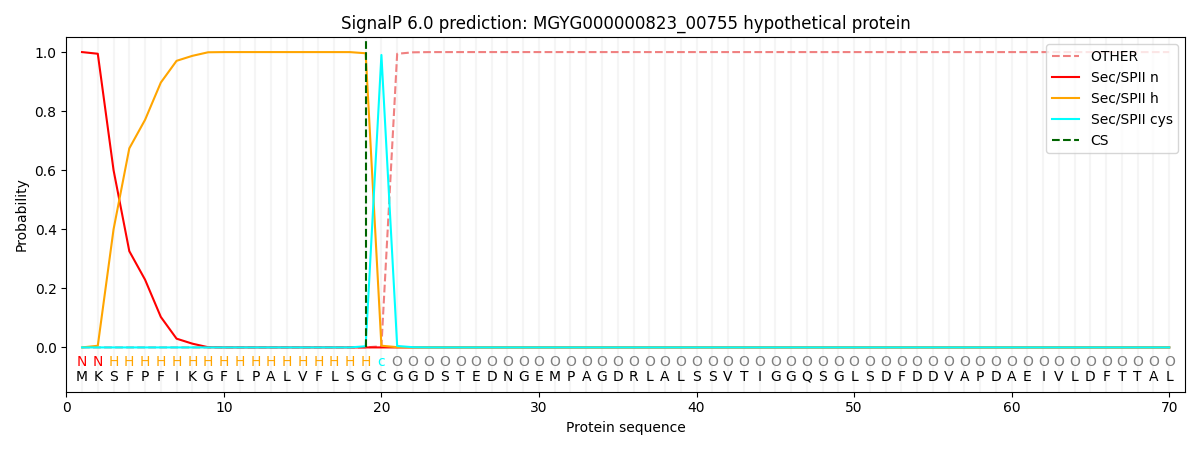
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000054 | 0.000000 | 0.000000 | 0.000000 |
