You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000824_00305
Basic Information
help
| Species |
CAG-485 sp900552315
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900552315
|
| CAZyme ID |
MGYG000000824_00305
|
| CAZy Family |
CBM74 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 522 |
|
56078.01 |
3.991 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000824 |
2120674 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 20555;
End: 22123
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| CBM74 |
21 |
333 |
2e-60 |
0.9935897435897436 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam16738
|
CBM26 |
3.34e-10 |
393 |
459 |
1 |
68 |
Starch-binding module 26. CBM26 is a carbohydrate-binding module that binds starch. |
This protein is predicted as OTHER
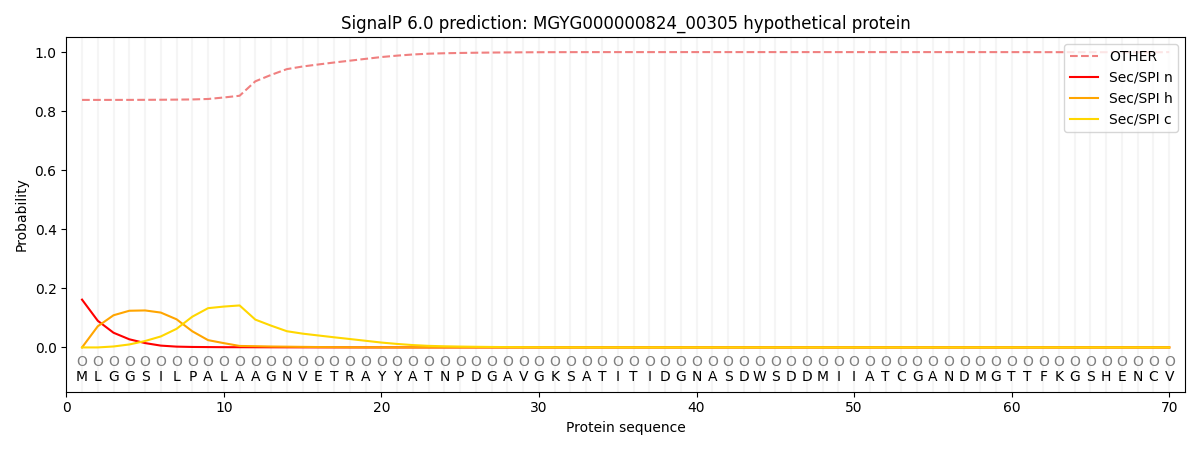
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.844532
|
0.154937
|
0.000118
|
0.000240
|
0.000088
|
0.000103
|
There is no transmembrane helices in MGYG000000824_00305.

