You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000825_00184
You are here: Home > Sequence: MGYG000000825_00184
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900544535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900544535 | |||||||||||
| CAZyme ID | MGYG000000825_00184 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | Acetyl esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45572; End: 47845 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 367 | 724 | 3.6e-57 | 0.9538461538461539 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 9.26e-50 | 339 | 666 | 83 | 421 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| COG0657 | Aes | 1.84e-35 | 12 | 331 | 8 | 312 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 2.07e-20 | 95 | 179 | 1 | 86 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| pfam00295 | Glyco_hydro_28 | 5.39e-18 | 373 | 721 | 9 | 319 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PRK10162 | PRK10162 | 4.87e-17 | 72 | 178 | 64 | 168 | acetyl esterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP73449.1 | 0.0 | 1 | 757 | 3 | 759 |
| QCD39802.1 | 0.0 | 1 | 757 | 3 | 759 |
| QCD43023.1 | 0.0 | 12 | 751 | 12 | 753 |
| AKQ46012.1 | 4.11e-160 | 336 | 728 | 19 | 410 |
| ARS40669.1 | 8.97e-159 | 336 | 728 | 19 | 410 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 1.05e-33 | 339 | 708 | 45 | 451 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 4.19e-30 | 341 | 734 | 30 | 439 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 1QZ3_A | 2.07e-17 | 77 | 183 | 58 | 166 | CRYSTALSTRUCTURE OF MUTANT M211S/R215L OF CARBOXYLESTERASE EST2 COMPLEXED WITH HEXADECANESULFONATE [Alicyclobacillus acidocaldarius],1U4N_A Crystal Structure Analysis of the M211S/R215L EST2 mutant [Alicyclobacillus acidocaldarius] |
| 4MXN_A | 2.44e-17 | 339 | 550 | 22 | 215 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 1EVQ_A | 3.75e-17 | 77 | 183 | 58 | 166 | THECRYSTAL STRUCTURE OF THE THERMOPHILIC CARBOXYLESTERASE EST2 FROM ALICYCLOBACILLUS ACIDOCALDARIUS [Alicyclobacillus acidocaldarius] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q50681 | 6.95e-19 | 58 | 301 | 143 | 375 | Probable carboxylic ester hydrolase LipM OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipM PE=1 SV=1 |
| Q949Z1 | 1.68e-17 | 339 | 639 | 80 | 363 | Polygalacturonase At1g48100 OS=Arabidopsis thaliana OX=3702 GN=At1g48100 PE=2 SV=1 |
| P15922 | 8.53e-17 | 341 | 729 | 154 | 598 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| I6Y9F7 | 1.55e-16 | 67 | 267 | 141 | 335 | Esterase LipQ OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipQ PE=1 SV=1 |
| P96402 | 5.68e-16 | 81 | 272 | 143 | 330 | Esterase LipC OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
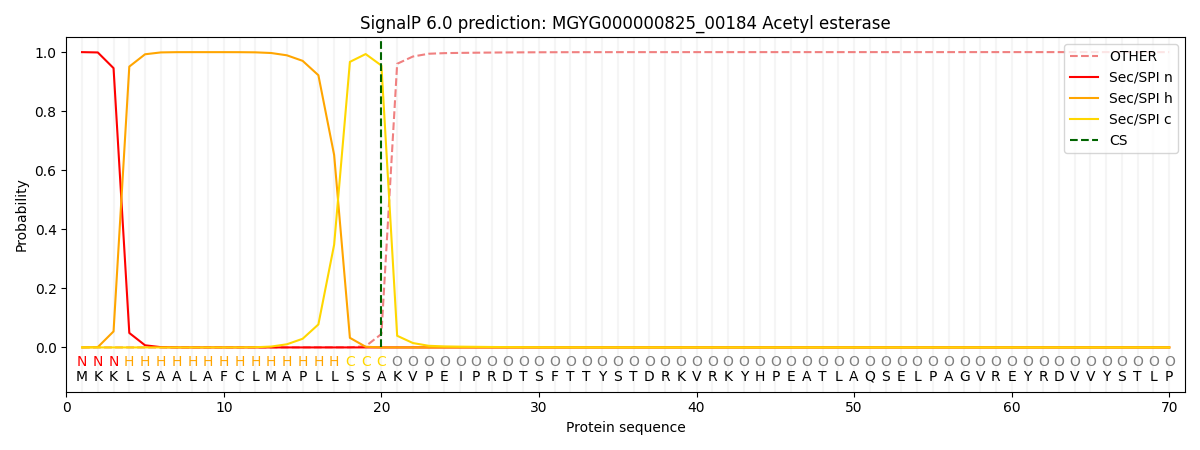
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000414 | 0.998602 | 0.000419 | 0.000184 | 0.000180 | 0.000165 |
