You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000825_00967
You are here: Home > Sequence: MGYG000000825_00967
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900544535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900544535 | |||||||||||
| CAZyme ID | MGYG000000825_00967 | |||||||||||
| CAZy Family | GT92 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 77703; End: 78578 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT92 | 52 | 273 | 1.4e-28 | 0.8207885304659498 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01697 | Glyco_transf_92 | 1.68e-29 | 63 | 212 | 19 | 183 | Glycosyltransferase family 92. Members of this family act as galactosyltransferases, belonging to glycosyltransferase family 92. The aligned region contains several conserved cysteine residues and several charged residues that may be catalytic residues. This is supported by the inclusion of this family in the GT-A glycosyl transferase superfamily. |
| pfam13704 | Glyco_tranf_2_4 | 1.42e-13 | 58 | 142 | 1 | 85 | Glycosyl transferase family 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD39434.1 | 1.67e-192 | 1 | 291 | 1 | 291 |
| QCP73126.1 | 1.67e-192 | 1 | 291 | 1 | 291 |
| ADY36745.1 | 8.29e-142 | 1 | 291 | 1 | 290 |
| SCD19281.1 | 6.95e-134 | 1 | 289 | 31 | 319 |
| QPH38826.1 | 9.24e-133 | 3 | 291 | 37 | 325 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B9S2H4 | 5.73e-12 | 48 | 211 | 294 | 465 | Glycosyltransferase family 92 protein RCOM_0530710 OS=Ricinus communis OX=3988 GN=RCOM_0699480 PE=3 SV=1 |
| Q6YRM6 | 3.37e-11 | 51 | 160 | 316 | 426 | Glycosyltransferase family 92 protein Os08g0121900 OS=Oryza sativa subsp. japonica OX=39947 GN=Os08g0121900 PE=2 SV=1 |
| B9SLR1 | 1.91e-10 | 51 | 140 | 281 | 371 | Glycosyltransferase family 92 protein RCOM_0530710 OS=Ricinus communis OX=3988 GN=RCOM_0530710 PE=3 SV=1 |
| Q94K98 | 1.95e-10 | 41 | 140 | 283 | 383 | Glycosyltransferase family 92 protein At1g27200 OS=Arabidopsis thaliana OX=3702 GN=At1g27200 PE=2 SV=2 |
| Q9LTZ9 | 2.00e-08 | 65 | 223 | 275 | 447 | Galactan beta-1,4-galactosyltransferase GALS2 OS=Arabidopsis thaliana OX=3702 GN=GALS2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
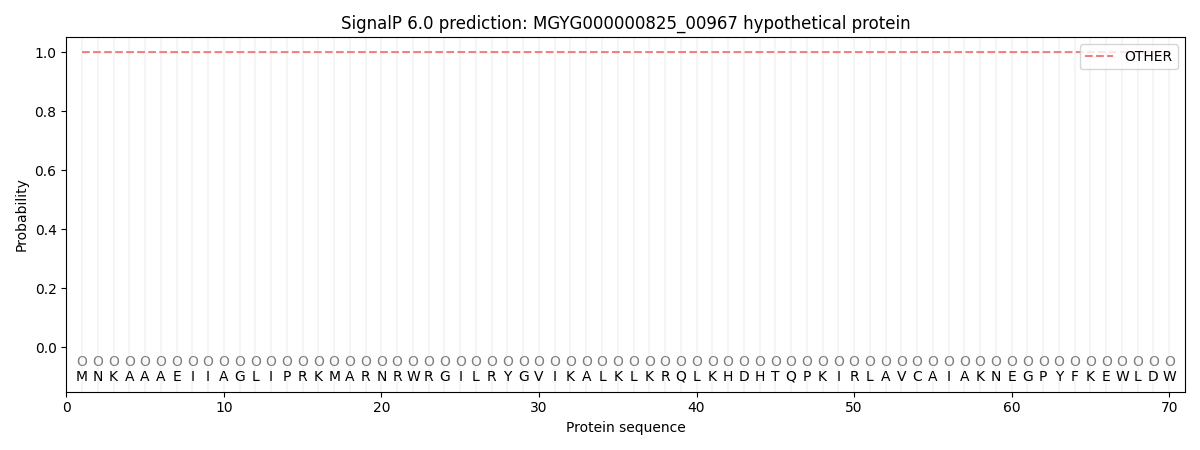
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000045 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
