You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000825_02066
You are here: Home > Sequence: MGYG000000825_02066
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900544535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900544535 | |||||||||||
| CAZyme ID | MGYG000000825_02066 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32374; End: 35361 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 107 | 292 | 3e-76 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09471 | Peptidase_M64 | 6.03e-100 | 732 | 987 | 1 | 259 | IgA Peptidase M64. This is a family of highly selective metallo-endopeptidases. The primary structure of the Clostridium ramosum IgA proteinase shows no significant overall similarity to any other known metallo-endopeptidase. |
| pfam16217 | M64_N | 3.35e-44 | 592 | 704 | 4 | 115 | Peptidase M64 N-terminus. This domain is found at the N-terminus of IgA Peptidase M64. Its function is unknown. |
| COG3866 | PelB | 3.05e-05 | 66 | 250 | 48 | 236 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.00e-04 | 100 | 292 | 3 | 186 | Amb_all domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD39200.1 | 5.20e-180 | 53 | 574 | 26 | 565 |
| QCP72891.1 | 5.20e-180 | 53 | 574 | 26 | 565 |
| QUT75603.1 | 1.13e-164 | 44 | 586 | 6 | 512 |
| QUT77823.1 | 7.83e-162 | 53 | 568 | 51 | 520 |
| QUT93111.1 | 3.32e-159 | 53 | 563 | 43 | 511 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4DF9_A | 9.17e-166 | 585 | 992 | 2 | 408 | Crystalstructure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343],4DF9_B Crystal structure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343],4DF9_C Crystal structure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343],4DF9_D Crystal structure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343],4DF9_E Crystal structure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343],4DF9_F Crystal structure of a putative peptidase (BF3526) from Bacteroides fragilis NCTC 9343 at 2.17 A resolution [Bacteroides fragilis NCTC 9343] |
| 3P1V_A | 2.32e-162 | 587 | 990 | 3 | 405 | Crystalstructure of a metallo-endopeptidases (BACOVA_00663) from Bacteroides ovatus at 1.93 A resolution [Bacteroides ovatus ATCC 8483],3P1V_B Crystal structure of a metallo-endopeptidases (BACOVA_00663) from Bacteroides ovatus at 1.93 A resolution [Bacteroides ovatus ATCC 8483] |
| 6FI2_A | 1.18e-07 | 60 | 215 | 46 | 193 | VexL:A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides [Achromobacter denitrificans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 1.86e-32 | 52 | 563 | 19 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 6.14e-32 | 52 | 563 | 19 | 414 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q4WL88 | 1.53e-31 | 52 | 563 | 20 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| B0XMA2 | 2.06e-31 | 52 | 563 | 20 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q5B297 | 2.10e-31 | 43 | 565 | 10 | 413 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
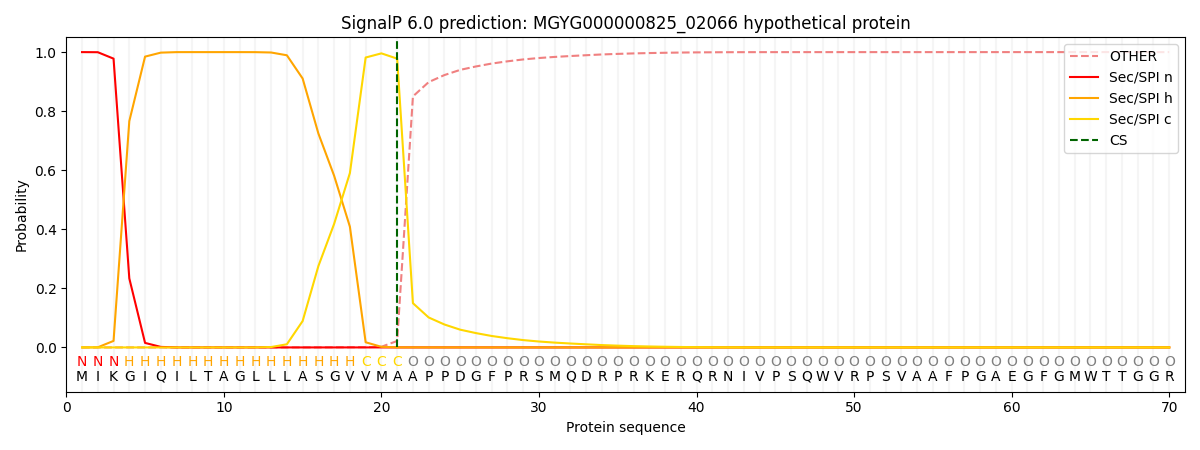
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000322 | 0.998924 | 0.000196 | 0.000182 | 0.000166 | 0.000165 |
