You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000825_02109
You are here: Home > Sequence: MGYG000000825_02109
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900544535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900544535 | |||||||||||
| CAZyme ID | MGYG000000825_02109 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16870; End: 19881 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 623 | 940 | 3.5e-93 | 0.9702970297029703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 6.44e-106 | 619 | 939 | 1 | 308 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.31e-94 | 664 | 937 | 1 | 261 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.09e-74 | 610 | 937 | 14 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| PRK15319 | PRK15319 | 0.007 | 214 | 535 | 809 | 1083 | fibronectin-binding autotransporter adhesin ShdA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05746.1 | 4.67e-104 | 676 | 951 | 7 | 284 |
| ADX05712.1 | 1.95e-71 | 617 | 918 | 70 | 380 |
| AUG58840.1 | 1.71e-67 | 617 | 939 | 275 | 591 |
| AEV69016.1 | 3.47e-66 | 617 | 936 | 472 | 786 |
| QJQ84452.1 | 5.70e-66 | 623 | 939 | 39 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XYZ_A | 1.23e-70 | 619 | 936 | 28 | 337 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6FHE_A | 3.93e-59 | 630 | 939 | 23 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 1VBR_A | 3.94e-59 | 616 | 952 | 3 | 326 | Crystalstructure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
| 3WUF_A | 9.01e-59 | 632 | 943 | 8 | 312 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3NIY_A | 1.46e-58 | 616 | 939 | 19 | 332 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 2.74e-65 | 619 | 936 | 518 | 827 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q60041 | 3.23e-57 | 616 | 952 | 22 | 345 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| P23360 | 3.75e-57 | 650 | 936 | 61 | 320 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| A1CHQ0 | 5.00e-57 | 648 | 943 | 57 | 317 | Probable endo-1,4-beta-xylanase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=xlnC PE=2 SV=1 |
| Q0H904 | 1.44e-55 | 650 | 943 | 59 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
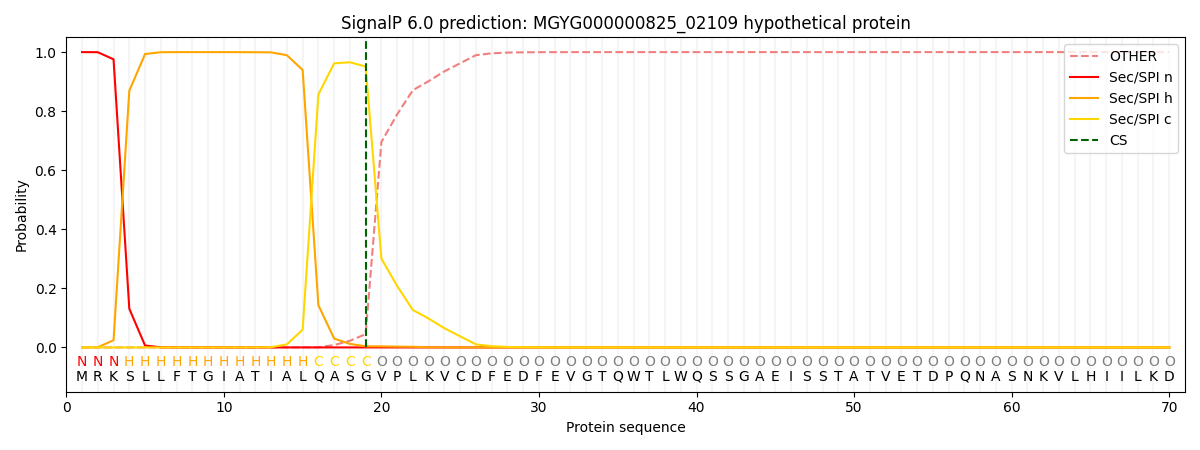
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000377 | 0.998922 | 0.000208 | 0.000162 | 0.000145 | 0.000136 |
