You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000839_02039
You are here: Home > Sequence: MGYG000000839_02039
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
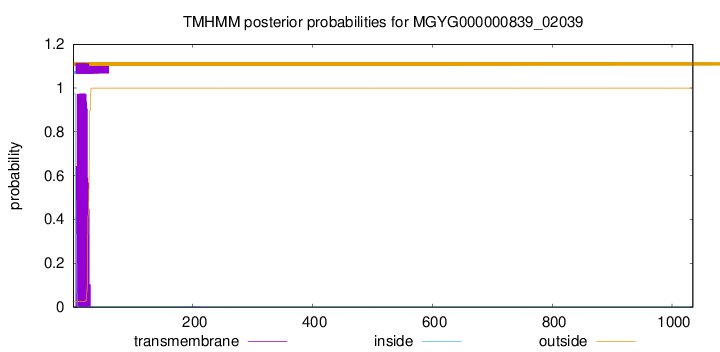
TMHMM annotations
Basic Information help
| Species | Clostridium sp900540255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900540255 | |||||||||||
| CAZyme ID | MGYG000000839_02039 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13262; End: 16369 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 2.66e-23 | 632 | 910 | 24 | 323 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 1.82e-22 | 679 | 956 | 26 | 323 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 2.45e-21 | 768 | 1002 | 26 | 323 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG4193 | LytD | 1.46e-19 | 337 | 501 | 50 | 223 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5492 | YjdB | 7.20e-18 | 798 | 939 | 11 | 157 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAS62016.1 | 1.34e-147 | 132 | 628 | 74 | 583 |
| AYE33874.1 | 1.34e-147 | 132 | 628 | 74 | 583 |
| ASW42466.1 | 1.71e-140 | 133 | 610 | 131 | 669 |
| QBJ74559.1 | 1.28e-139 | 132 | 610 | 74 | 566 |
| SLK13348.1 | 1.28e-139 | 132 | 610 | 74 | 566 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 1.86e-27 | 210 | 500 | 29 | 276 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
| 4PI7_A | 2.27e-06 | 358 | 501 | 55 | 206 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.85e-25 | 210 | 507 | 397 | 651 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.20e-25 | 210 | 507 | 441 | 695 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
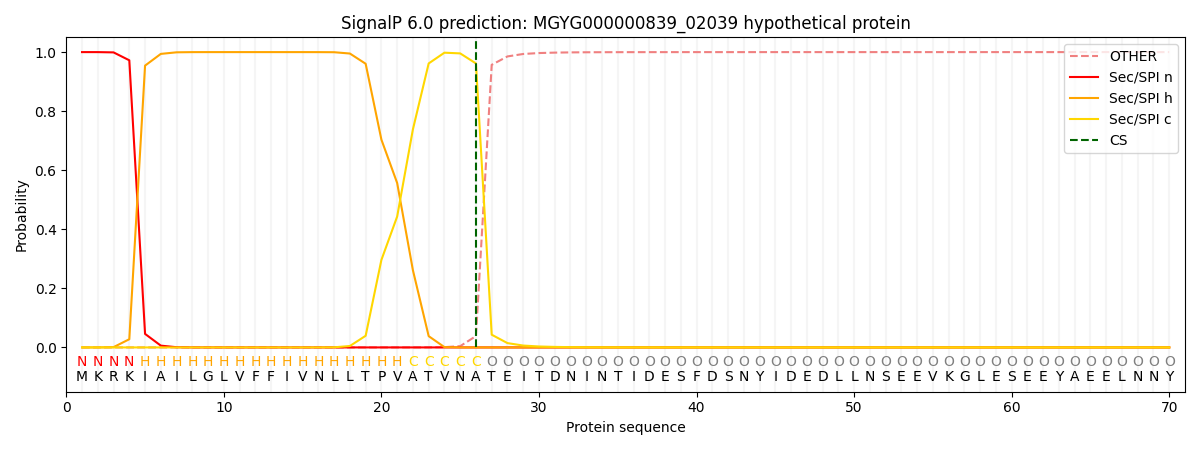
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000245 | 0.999105 | 0.000167 | 0.000165 | 0.000147 | 0.000139 |