You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000851_00445
You are here: Home > Sequence: MGYG000000851_00445
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA7173 sp900552325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; UBA7173 sp900552325 | |||||||||||
| CAZyme ID | MGYG000000851_00445 | |||||||||||
| CAZy Family | GH87 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1177; End: 5151 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH87 | 40 | 585 | 5.8e-125 | 0.8023450586264657 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14490 | CBM6-CBM35-CBM36_like_1 | 5.41e-29 | 42 | 211 | 1 | 156 | uncharacterized members of the carbohydrate binding module 6 (CBM6) and CBM35_like superfamily. Carbohydrate binding module family 6 (CBM6, family 6 CBM), also known as cellulose binding domain family VI (CBD VI), and related CBMs (CBM35 and CBM36). These are non-catalytic carbohydrate binding domains found in a range of enzymes that display activities against a diverse range of carbohydrate targets, including mannan, xylan, beta-glucans, cellulose, agarose, and arabinans. These domains facilitate the strong binding of the appended catalytic modules to their dedicated, insoluble substrates. Many of these CBMs are associated with glycoside hydrolase (GH) domains. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. CBM36s are calcium-dependent xylan binding domains. CBM35s display conserved specificity through extensive sequence similarity, but divergent function through their appended catalytic modules. |
| pfam13229 | Beta_helix | 2.90e-07 | 393 | 572 | 8 | 155 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam12708 | Pectate_lyase_3 | 1.26e-04 | 230 | 309 | 2 | 72 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG5434 | Pgu1 | 0.003 | 228 | 272 | 81 | 127 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH89883.1 | 4.95e-210 | 10 | 930 | 7 | 878 |
| ANR73273.1 | 6.96e-210 | 2 | 945 | 3 | 896 |
| QUB45559.1 | 7.18e-210 | 2 | 945 | 4 | 897 |
| QUB58650.1 | 2.81e-209 | 10 | 930 | 8 | 879 |
| QUB56055.1 | 2.81e-209 | 10 | 930 | 8 | 879 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5ZRU_A | 2.65e-115 | 40 | 610 | 11 | 554 | ChainA, Alpha-1,3-glucanase [Niallia circulans],5ZRU_C Chain C, Alpha-1,3-glucanase [Niallia circulans] |
| 6K0M_A | 4.69e-22 | 41 | 512 | 13 | 434 | Catalyticdomain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 [Paenibacillus glycanilyticus],6K0N_A Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose [Paenibacillus glycanilyticus] |
| 6K0P_A | 4.45e-21 | 41 | 512 | 13 | 434 | Catalyticdomain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose [Paenibacillus glycanilyticus] |
| 6K0S_A | 4.45e-21 | 41 | 512 | 13 | 434 | Catalyticdomain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose [Paenibacillus glycanilyticus],6K0V_A Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides [Paenibacillus glycanilyticus],6K0V_B Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides [Paenibacillus glycanilyticus],6K0V_C Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides [Paenibacillus glycanilyticus],6K0V_D Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides [Paenibacillus glycanilyticus] |
| 6K0Q_A | 3.40e-17 | 41 | 512 | 13 | 434 | Catalyticdomain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose [Paenibacillus glycanilyticus],6K0U_A Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides [Paenibacillus glycanilyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
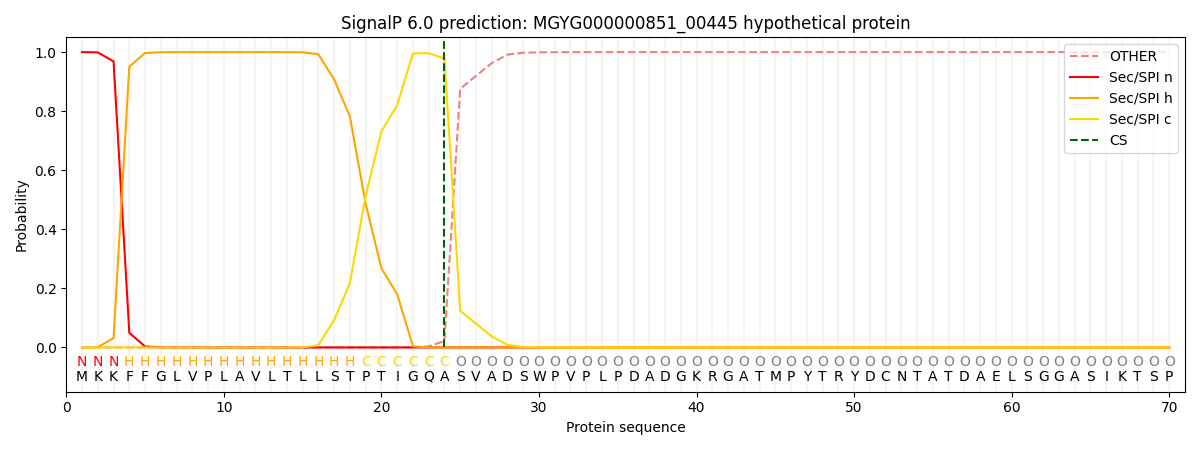
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999089 | 0.000183 | 0.000174 | 0.000158 | 0.000145 |
