You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000852_01815
You are here: Home > Sequence: MGYG000000852_01815
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900545525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900545525 | |||||||||||
| CAZyme ID | MGYG000000852_01815 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3517; End: 5364 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 251 | 545 | 1.8e-82 | 0.9893617021276596 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.98e-30 | 253 | 539 | 1 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.55e-23 | 247 | 568 | 49 | 390 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26200.1 | 3.15e-144 | 251 | 570 | 40 | 358 |
| ALJ47659.1 | 3.15e-144 | 251 | 570 | 40 | 358 |
| QUT31547.1 | 3.15e-144 | 251 | 570 | 40 | 358 |
| QRM99696.1 | 3.15e-144 | 251 | 570 | 40 | 358 |
| QDH53449.1 | 3.15e-144 | 251 | 570 | 40 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LX4_A | 3.34e-74 | 250 | 568 | 13 | 313 | CrystalStructure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_B Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_C Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_D Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],6R2J_A Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_B Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_C Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_D Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501] |
| 4EE9_A | 1.05e-72 | 247 | 568 | 1 | 306 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 2.91e-72 | 247 | 568 | 1 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 3.96e-72 | 247 | 568 | 1 | 306 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 4.08e-72 | 247 | 568 | 1 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P58599 | 3.48e-44 | 252 | 568 | 119 | 422 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| P17974 | 1.69e-42 | 252 | 568 | 121 | 424 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| Q5BDU5 | 2.82e-35 | 247 | 552 | 20 | 306 | Endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglA PE=1 SV=1 |
| O74706 | 8.11e-35 | 243 | 553 | 27 | 316 | Endo-beta-1,4-glucanase B OS=Aspergillus niger OX=5061 GN=eglB PE=1 SV=1 |
| A2QPC3 | 8.11e-35 | 243 | 553 | 27 | 316 | Probable endo-beta-1,4-glucanase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
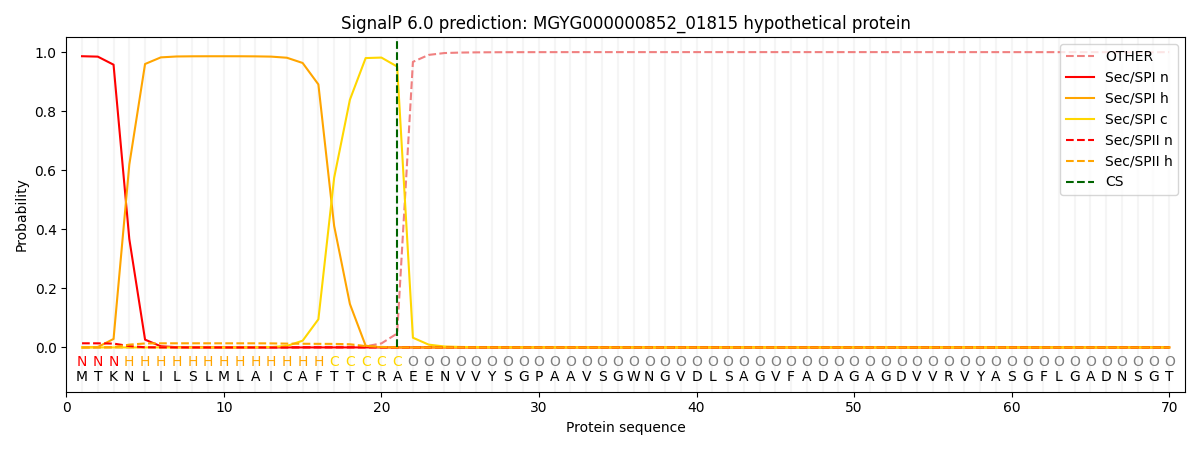
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000459 | 0.981629 | 0.017184 | 0.000253 | 0.000235 | 0.000222 |
