You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000853_00547
You are here: Home > Sequence: MGYG000000853_00547
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900549175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900549175 | |||||||||||
| CAZyme ID | MGYG000000853_00547 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25638; End: 27683 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 34 | 378 | 7.6e-86 | 0.990228013029316 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 8.25e-54 | 33 | 378 | 1 | 315 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 2.06e-32 | 27 | 378 | 30 | 339 | beta-galactosidase; Provisional |
| COG1874 | GanA | 2.40e-13 | 30 | 180 | 4 | 160 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 5.60e-04 | 54 | 180 | 8 | 137 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37835.1 | 0.0 | 1 | 677 | 1 | 672 |
| AGB28883.1 | 7.30e-287 | 18 | 678 | 337 | 1028 |
| ATC65137.1 | 4.57e-184 | 30 | 660 | 66 | 796 |
| ACB74649.1 | 4.38e-168 | 18 | 664 | 40 | 802 |
| QQA07701.1 | 9.00e-167 | 8 | 675 | 6 | 681 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W5F_A | 4.13e-29 | 26 | 477 | 7 | 398 | ChainA, Beta-galactosidase [Solanum lycopersicum],3W5F_B Chain B, Beta-galactosidase [Solanum lycopersicum],3W5G_A Chain A, Beta-galactosidase [Solanum lycopersicum],3W5G_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK5_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK5_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
| 6IK6_A | 9.69e-29 | 26 | 477 | 7 | 398 | ChainA, Beta-galactosidase [Solanum lycopersicum],6IK6_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK7_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK7_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK8_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK8_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
| 4E8C_A | 2.34e-25 | 36 | 377 | 12 | 325 | Crystalstructure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8C_B Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8D_A Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4],4E8D_B Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4] |
| 7KDV_A | 6.35e-25 | 37 | 377 | 28 | 337 | ChainA, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
| 5GSL_A | 1.22e-24 | 27 | 180 | 6 | 181 | Glycosidehydrolase A [Pyrococcus horikoshii OT3],5GSL_B Glycoside hydrolase A [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GX69 | 2.35e-31 | 20 | 378 | 18 | 335 | Beta-galactosidase 16 OS=Arabidopsis thaliana OX=3702 GN=BGAL16 PE=2 SV=2 |
| Q9MAJ7 | 2.50e-31 | 3 | 480 | 8 | 424 | Beta-galactosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BGAL5 PE=2 SV=1 |
| Q9FFN4 | 3.17e-31 | 27 | 456 | 32 | 440 | Beta-galactosidase 6 OS=Arabidopsis thaliana OX=3702 GN=BGAL6 PE=2 SV=1 |
| Q9SCV9 | 1.40e-30 | 4 | 428 | 11 | 386 | Beta-galactosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BGAL3 PE=2 SV=1 |
| Q6Z6K4 | 1.84e-30 | 19 | 464 | 30 | 416 | Beta-galactosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=Os02g0219200 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
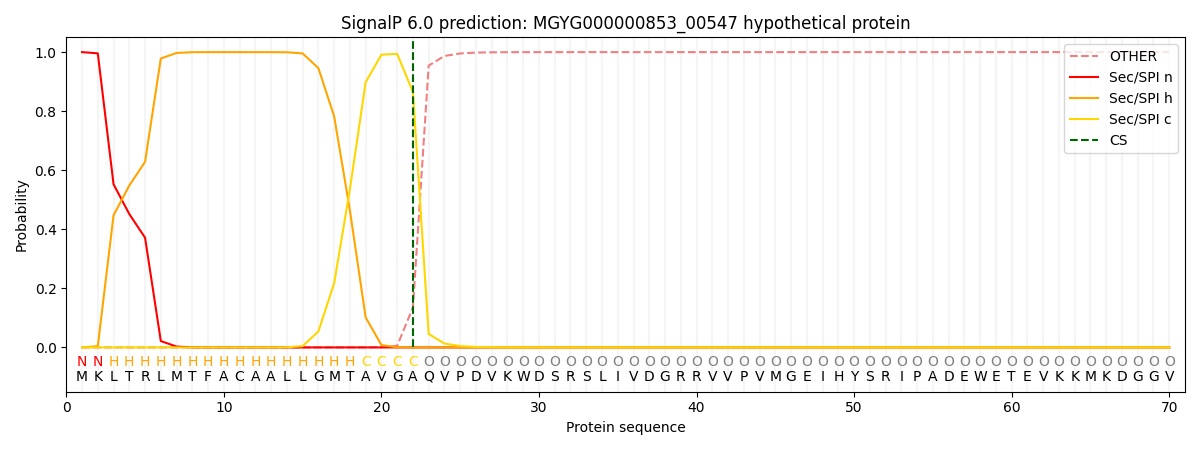
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999079 | 0.000205 | 0.000165 | 0.000140 | 0.000134 |
