You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000853_01290
Basic Information
help
| Species |
Prevotella sp900549175
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900549175
|
| CAZyme ID |
MGYG000000853_01290
|
| CAZy Family |
GH28 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 479 |
|
53619.92 |
7.9113 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000853 |
3157276 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 12405;
End: 13844
Strand: -
|
No EC number prediction in MGYG000000853_01290.
| Family |
Start |
End |
Evalue |
family coverage |
| GH28 |
189 |
445 |
1.2e-27 |
0.76 |
MGYG000000853_01290 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6NZS_A
|
2.68e-06 |
124 |
417 |
199 |
514 |
DextranaseAoDex KQ11 [Pseudarthrobacter oxydans] |
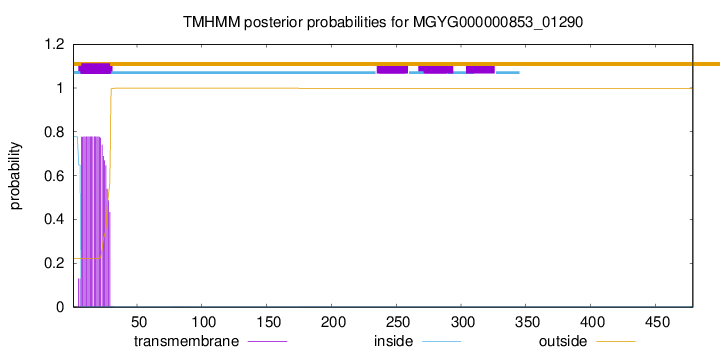
This protein is predicted as SP
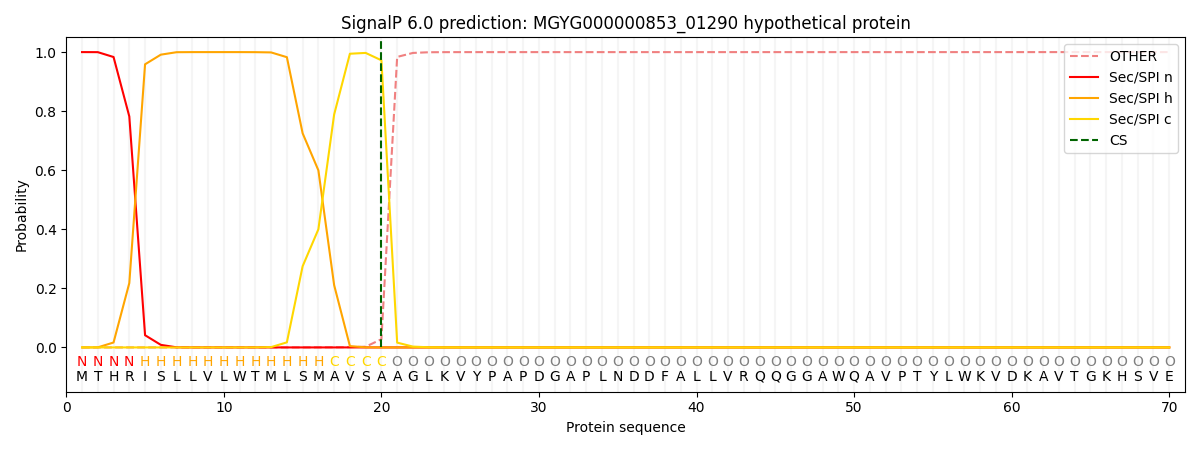
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000285
|
0.999025
|
0.000166
|
0.000177
|
0.000168
|
0.000154
|