You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000853_01546
You are here: Home > Sequence: MGYG000000853_01546
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900549175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900549175 | |||||||||||
| CAZyme ID | MGYG000000853_01546 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5428; End: 7767 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 465 | 723 | 1.4e-47 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.91e-32 | 467 | 723 | 98 | 309 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.39e-30 | 467 | 722 | 56 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 8.61e-20 | 472 | 720 | 126 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.36e-14 | 56 | 135 | 12 | 90 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 4.51e-10 | 88 | 144 | 1 | 56 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQZ02683.1 | 1.58e-288 | 18 | 775 | 18 | 773 |
| ADE83221.1 | 3.26e-175 | 1 | 777 | 1 | 464 |
| AXH21505.1 | 8.29e-172 | 23 | 725 | 29 | 742 |
| EDV05072.1 | 8.29e-172 | 23 | 725 | 29 | 742 |
| QRX63290.1 | 3.70e-166 | 7 | 725 | 15 | 728 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6LPS_A | 7.07e-15 | 454 | 719 | 108 | 346 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 6FHE_A | 8.38e-15 | 454 | 723 | 111 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 4MGS_A | 8.75e-15 | 149 | 288 | 2 | 144 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 9.76e-15 | 185 | 288 | 35 | 138 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 1W32_A | 1.23e-14 | 486 | 724 | 118 | 343 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O59859 | 5.41e-14 | 456 | 724 | 115 | 325 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
| P07528 | 2.37e-13 | 454 | 719 | 153 | 391 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P40943 | 3.39e-13 | 459 | 642 | 150 | 298 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 PE=1 SV=1 |
| O69230 | 4.24e-13 | 457 | 667 | 467 | 648 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P14768 | 8.78e-13 | 486 | 724 | 381 | 606 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
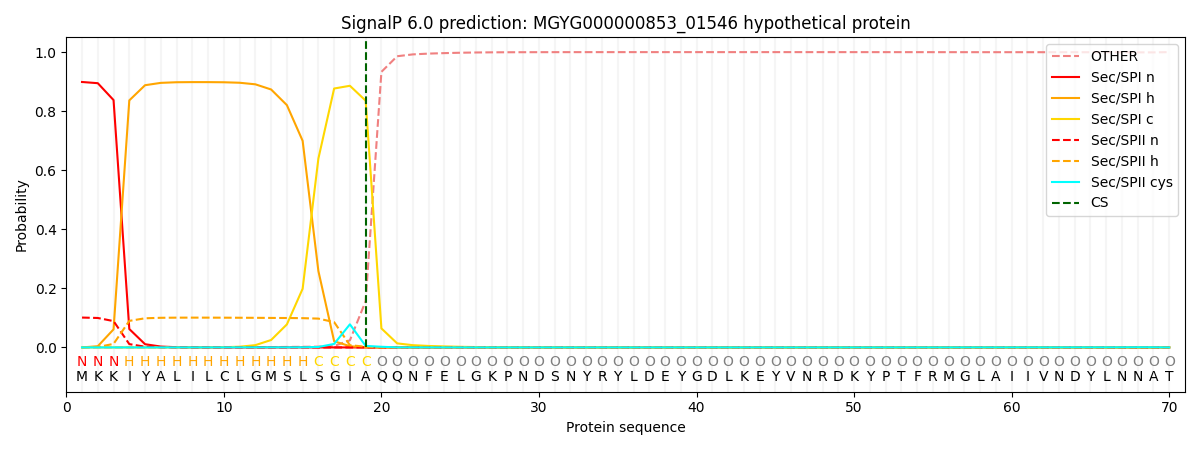
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001055 | 0.893341 | 0.104790 | 0.000259 | 0.000268 | 0.000247 |
