You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000853_01612
You are here: Home > Sequence: MGYG000000853_01612
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900549175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900549175 | |||||||||||
| CAZyme ID | MGYG000000853_01612 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6717; End: 8123 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 34 | 316 | 4.9e-32 | 0.8715277777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 7.12e-18 | 34 | 351 | 96 | 405 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PRK10531 | PRK10531 | 4.62e-11 | 29 | 261 | 92 | 350 | putative acyl-CoA thioester hydrolase. |
| PLN02488 | PLN02488 | 2.47e-09 | 13 | 348 | 186 | 492 | probable pectinesterase/pectinesterase inhibitor |
| pfam01095 | Pectinesterase | 2.94e-09 | 26 | 349 | 1 | 296 | Pectinesterase. |
| PLN02995 | PLN02995 | 6.45e-09 | 18 | 302 | 211 | 487 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP72097.1 | 7.09e-217 | 7 | 444 | 6 | 443 |
| QCD38407.1 | 7.09e-217 | 7 | 444 | 6 | 443 |
| QNT65238.1 | 1.03e-135 | 16 | 425 | 657 | 1072 |
| QCD41145.1 | 1.07e-129 | 22 | 433 | 1035 | 1443 |
| AEF12641.1 | 1.12e-98 | 25 | 440 | 1042 | 1459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 5.62e-11 | 164 | 353 | 140 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 2.37e-08 | 164 | 353 | 140 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 2.37e-08 | 164 | 353 | 140 | 339 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A8 | 1.43e-07 | 164 | 353 | 164 | 363 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| P0C1A9 | 1.43e-07 | 164 | 353 | 164 | 363 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
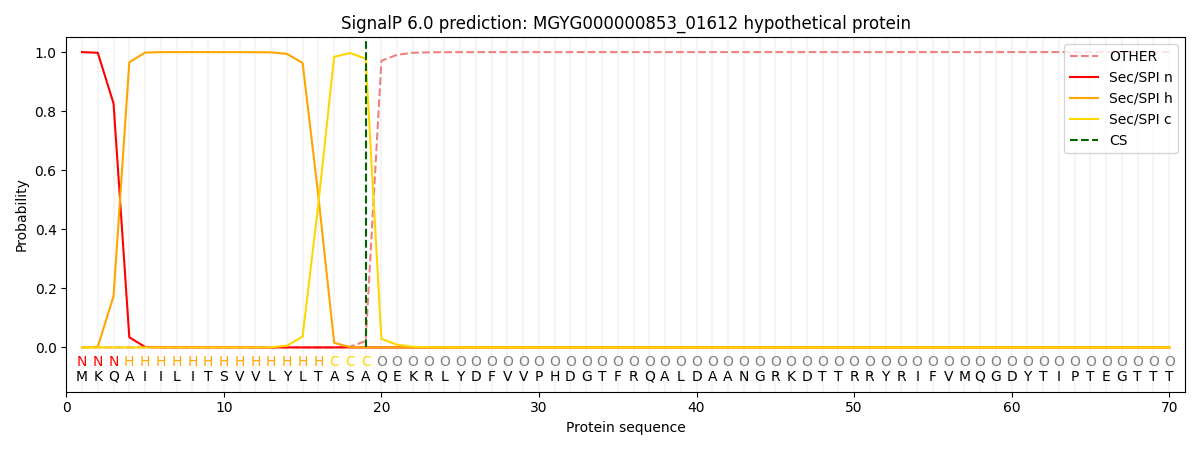
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000276 | 0.998912 | 0.000303 | 0.000162 | 0.000162 | 0.000147 |
