You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000853_01887
You are here: Home > Sequence: MGYG000000853_01887
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900549175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900549175 | |||||||||||
| CAZyme ID | MGYG000000853_01887 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5657; End: 7414 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 19 | 172 | 9.5e-22 | 0.6079295154185022 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 9.56e-34 | 17 | 250 | 42 | 312 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG4099 | COG4099 | 5.91e-10 | 24 | 169 | 176 | 332 | Predicted peptidase [General function prediction only]. |
| pfam17957 | Big_7 | 1.24e-08 | 254 | 327 | 1 | 67 | Bacterial Ig domain. This entry represents a bacterial ig-like domain that is found in glycosyl hydrolase enzymes. |
| pfam02230 | Abhydrolase_2 | 3.01e-07 | 36 | 169 | 14 | 173 | Phospholipase/Carboxylesterase. This family consists of both phospholipases and carboxylesterases with broad substrate specificity, and is structurally related to alpha/beta hydrolases pfam00561. |
| pfam10503 | Esterase_phd | 5.32e-07 | 23 | 143 | 2 | 134 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAV06734.1 | 9.28e-81 | 15 | 247 | 28 | 266 |
| ABG59303.1 | 9.50e-76 | 15 | 486 | 40 | 526 |
| ASB48751.1 | 5.11e-75 | 13 | 254 | 29 | 272 |
| ACX75364.1 | 1.07e-67 | 12 | 312 | 27 | 315 |
| ADL27269.1 | 1.07e-67 | 12 | 312 | 27 | 315 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WYD_A | 3.22e-14 | 19 | 183 | 18 | 183 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 3PDD_A | 2.87e-07 | 252 | 348 | 92 | 179 | ChainA, Glycoside hydrolase, family 9 [Acetivibrio thermocellus ATCC 27405] |
| 3PE9_A | 2.94e-07 | 254 | 348 | 2 | 87 | ChainA, Fibronectin(III)-like module [Acetivibrio thermocellus],3PE9_B Chain B, Fibronectin(III)-like module [Acetivibrio thermocellus],3PE9_C Chain C, Fibronectin(III)-like module [Acetivibrio thermocellus],3PE9_D Chain D, Fibronectin(III)-like module [Acetivibrio thermocellus] |
| 3DOH_A | 6.62e-07 | 19 | 167 | 155 | 323 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8YG19 | 8.60e-51 | 19 | 251 | 55 | 286 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
| A2QYU7 | 1.03e-13 | 22 | 225 | 43 | 264 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
| Q7RWX8 | 6.35e-12 | 16 | 246 | 46 | 290 | Feruloyl esterase D OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=faeD-3.544 PE=1 SV=1 |
| Q9Y871 | 3.04e-11 | 37 | 247 | 309 | 535 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
| A1CC33 | 5.49e-11 | 22 | 225 | 45 | 266 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
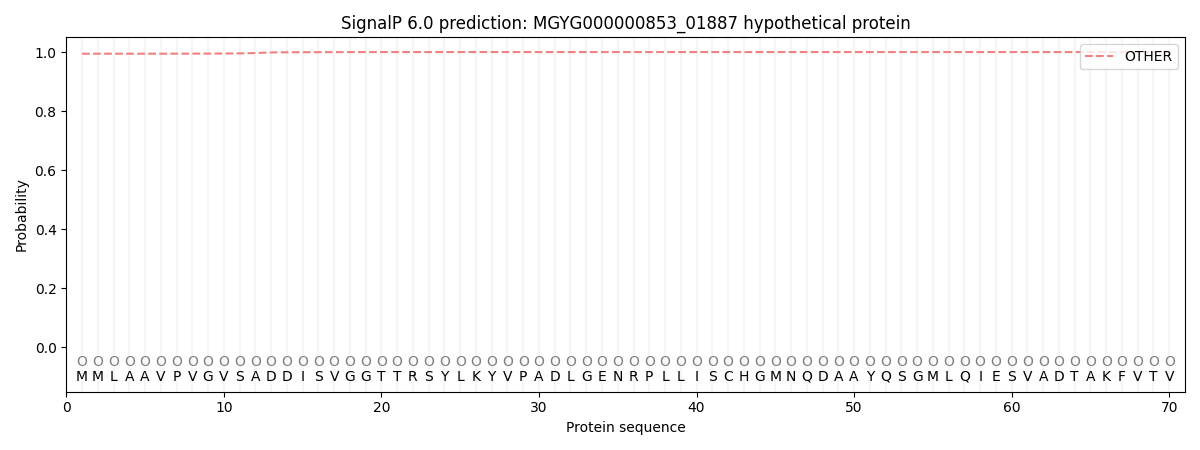
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.995090 | 0.004921 | 0.000006 | 0.000006 | 0.000003 | 0.000005 |
