You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000854_01157
You are here: Home > Sequence: MGYG000000854_01157
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900548345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900548345 | |||||||||||
| CAZyme ID | MGYG000000854_01157 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29323; End: 32919 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 16 | 411 | 2.3e-90 | 0.9653179190751445 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01120 | Alpha_L_fucos | 8.18e-99 | 30 | 407 | 4 | 333 | Alpha-L-fucosidase. |
| smart00812 | Alpha_L_fucos | 4.84e-98 | 26 | 468 | 1 | 378 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| COG3669 | AfuC | 3.07e-70 | 58 | 523 | 1 | 424 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| pfam16405 | DUF5013 | 6.08e-13 | 629 | 769 | 4 | 145 | Domain of unknown function (DUF5013). This small family of proteins is functionally uncharacterized. This family is found in various Bacteroides and Parabacteroides species. Proteins in this family are around 400 amino acids in length. |
| pfam16757 | Fucosidase_C | 1.40e-07 | 440 | 523 | 1 | 89 | Alpha-L-fucosidase C-terminal domain. The C-terminal domain of Structure 1hl8 is constructed of eight anti-parallel-strands packed into two-sheets of five and three strands, respectively, forming a two-layer-sandwich containing a Greek key motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB29378.1 | 9.24e-280 | 19 | 789 | 86 | 859 |
| QCD36457.1 | 5.43e-185 | 26 | 523 | 32 | 544 |
| VTR48024.1 | 2.52e-183 | 28 | 523 | 35 | 547 |
| ADY36480.1 | 2.60e-183 | 26 | 523 | 36 | 548 |
| QDO68463.1 | 3.31e-183 | 24 | 523 | 30 | 544 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LJJ_A | 5.11e-69 | 32 | 531 | 43 | 516 | ChainA, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135],7LK7_A Chain A, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135] |
| 7LNP_A | 2.38e-68 | 32 | 531 | 43 | 516 | ChainA, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135],7LNP_C Chain C, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135],7LNP_D Chain D, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135],7LNP_E Chain E, Exo-alpha-L-galactosidase [Phocaeicola plebeius DSM 17135] |
| 4NI3_A | 1.92e-49 | 29 | 530 | 4 | 496 | ChainA, Alpha-fucosidase GH29 [Fusarium graminearum],4NI3_B Chain B, Alpha-fucosidase GH29 [Fusarium graminearum],4PSP_A Chain A, Alpha-fucosidase GH29 [Fusarium graminearum],4PSP_B Chain B, Alpha-fucosidase GH29 [Fusarium graminearum],4PSR_A Chain A, Alpha-fucosidase GH29 [Fusarium graminearum],4PSR_B Chain B, Alpha-fucosidase GH29 [Fusarium graminearum] |
| 1HL8_A | 4.28e-36 | 32 | 510 | 8 | 431 | CrystalStructure Of Thermotoga Maritima Alpha-Fucosidase [Thermotoga maritima MSB8],1HL8_B Crystal Structure Of Thermotoga Maritima Alpha-Fucosidase [Thermotoga maritima MSB8],1HL9_A Crystal Structure Of Thermotoga Maritima Alpha-Fucosidase In Complex With A Mechanism Based Inhibitor [Thermotoga maritima MSB8],1HL9_B Crystal Structure Of Thermotoga Maritima Alpha-Fucosidase In Complex With A Mechanism Based Inhibitor [Thermotoga maritima MSB8] |
| 1ODU_A | 5.77e-36 | 32 | 510 | 8 | 431 | CrystalStructure Of Thermotoga Maritima Alpha-Fucosidase In Complex With Fucose [Thermotoga maritima MSB8],1ODU_B Crystal Structure Of Thermotoga Maritima Alpha-Fucosidase In Complex With Fucose [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q99LJ1 | 4.87e-34 | 25 | 520 | 16 | 440 | Tissue alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca1 PE=1 SV=1 |
| P17164 | 1.40e-33 | 18 | 515 | 21 | 446 | Tissue alpha-L-fucosidase OS=Rattus norvegicus OX=10116 GN=Fuca1 PE=1 SV=1 |
| Q2KIM0 | 6.79e-33 | 32 | 492 | 40 | 426 | Tissue alpha-L-fucosidase OS=Bos taurus OX=9913 GN=FUCA1 PE=2 SV=1 |
| Q9BTY2 | 5.31e-32 | 32 | 510 | 34 | 446 | Plasma alpha-L-fucosidase OS=Homo sapiens OX=9606 GN=FUCA2 PE=1 SV=2 |
| P04066 | 1.71e-31 | 32 | 521 | 37 | 456 | Tissue alpha-L-fucosidase OS=Homo sapiens OX=9606 GN=FUCA1 PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
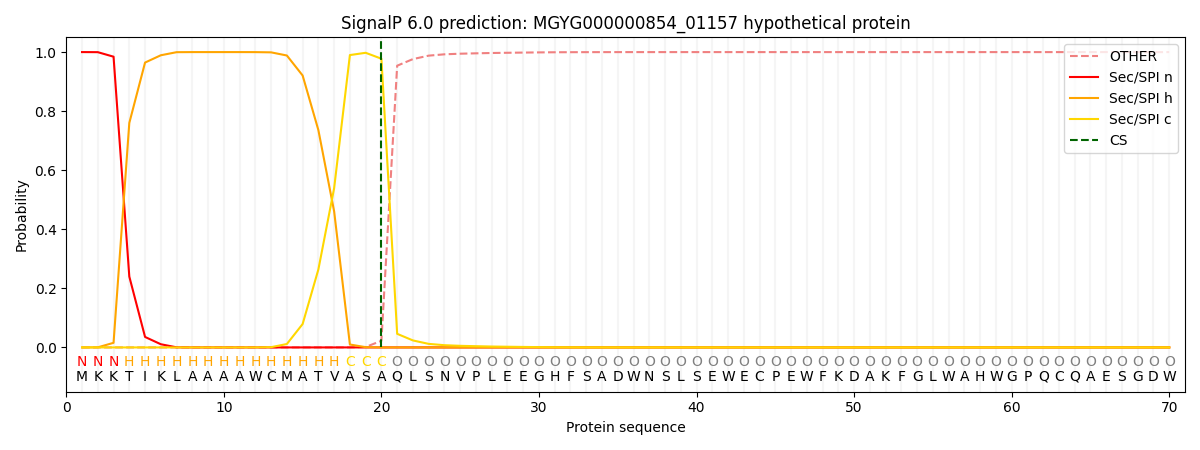
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000281 | 0.999112 | 0.000168 | 0.000160 | 0.000147 | 0.000141 |
