You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000859_02761
You are here: Home > Sequence: MGYG000000859_02761
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
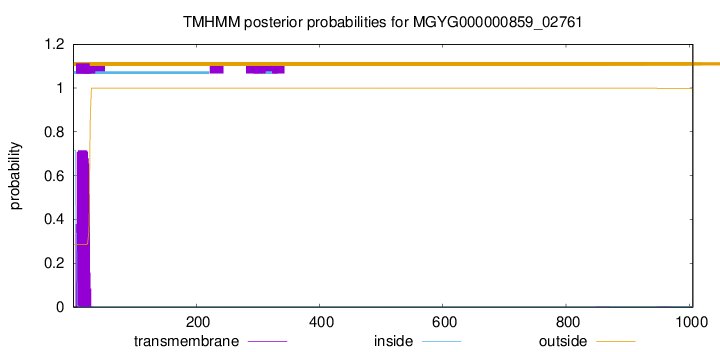
TMHMM annotations
Basic Information help
| Species | Faecalimonas sp900546325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas sp900546325 | |||||||||||
| CAZyme ID | MGYG000000859_02761 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4251; End: 7271 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08481 | GBS_Bsp-like | 1.05e-33 | 677 | 764 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 8.18e-33 | 480 | 567 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 5.55e-27 | 292 | 369 | 10 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 2.48e-25 | 580 | 662 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 4.72e-25 | 385 | 465 | 6 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEN97541.1 | 2.47e-158 | 191 | 872 | 820 | 1514 |
| QEI32622.1 | 4.15e-130 | 196 | 772 | 552 | 1127 |
| QRT31212.1 | 4.15e-130 | 196 | 772 | 552 | 1127 |
| QHB25110.1 | 4.15e-130 | 196 | 772 | 552 | 1127 |
| QRO35993.1 | 1.09e-129 | 190 | 846 | 591 | 1263 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
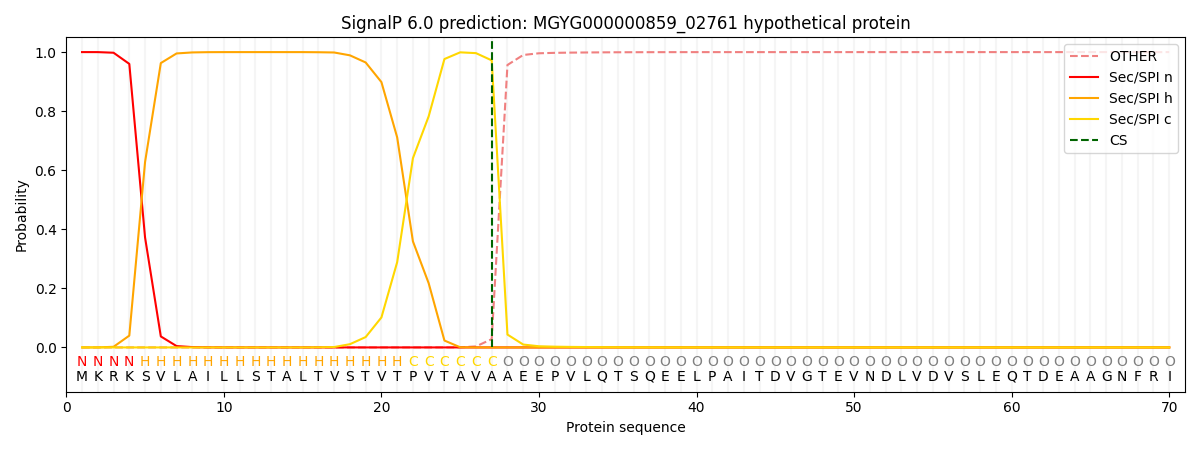
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000282 | 0.998980 | 0.000171 | 0.000220 | 0.000180 | 0.000155 |