You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000863_00390
You are here: Home > Sequence: MGYG000000863_00390
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1490 sp900547395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; UMGS1490; UMGS1490 sp900547395 | |||||||||||
| CAZyme ID | MGYG000000863_00390 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 130503; End: 134048 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 88 | 561 | 5.3e-69 | 0.5039893617021277 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 5.96e-26 | 142 | 533 | 66 | 435 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.71e-20 | 136 | 529 | 62 | 447 | beta-D-glucuronidase; Provisional |
| pfam07691 | PA14 | 2.78e-18 | 896 | 1021 | 4 | 130 | PA14 domain. This domain forms an insert in bacterial beta-glucosidases and is found in other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins, including anthrax protective antigen (PA). The domain also occurs in a Dictyostelium prespore-cell-inducing factor Psi and in fibrocystin, the mammalian protein whose mutation leads to polycystic kidney and hepatic disease. The crystal structure of PA shows that this domain (named PA14 after its location in the PA20 pro-peptide) has a beta-barrel structure. The PA14 domain sequence suggests a binding function, rather than a catalytic role. The PA14 domain distribution is compatible with carbohydrate binding. |
| smart00758 | PA14 | 9.10e-18 | 896 | 1021 | 2 | 126 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
| PRK10340 | ebgA | 7.52e-17 | 145 | 529 | 115 | 474 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88121.1 | 9.90e-213 | 12 | 1179 | 14 | 1130 |
| QDO68997.1 | 2.07e-212 | 22 | 1179 | 21 | 1128 |
| ALJ60861.1 | 7.67e-212 | 12 | 1179 | 14 | 1130 |
| QRQ50392.1 | 1.75e-210 | 22 | 1179 | 11 | 1118 |
| QUT46481.1 | 3.45e-210 | 13 | 1179 | 9 | 1130 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ED1_A | 1.12e-26 | 138 | 525 | 76 | 447 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
| 6ECA_A | 8.19e-21 | 108 | 523 | 70 | 463 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 6D1P_A | 2.93e-18 | 137 | 525 | 86 | 461 | Apostructure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii],6D1P_B Apo structure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii] |
| 3CMG_A | 1.61e-17 | 122 | 525 | 38 | 421 | Crystalstructure of putative beta-galactosidase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 5Z1A_A | 1.65e-17 | 122 | 525 | 57 | 440 | Thecrystal structure of Bacteroides fragilis beta-glucuronidase in complex with uronic isofagomine [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 7.29e-29 | 92 | 525 | 58 | 464 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| P77989 | 2.64e-23 | 136 | 525 | 53 | 410 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P26257 | 9.85e-20 | 164 | 525 | 78 | 411 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| T2KN75 | 1.91e-15 | 110 | 540 | 58 | 469 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| P06864 | 1.16e-13 | 145 | 529 | 115 | 474 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
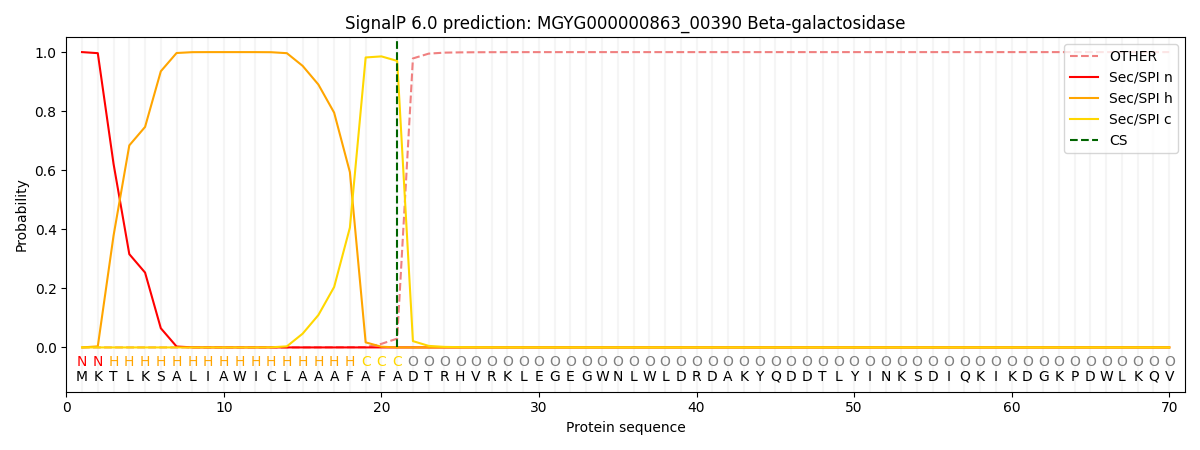
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000240 | 0.999122 | 0.000166 | 0.000159 | 0.000146 | 0.000134 |
