You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000870_02500
You are here: Home > Sequence: MGYG000000870_02500
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp004167605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp004167605 | |||||||||||
| CAZyme ID | MGYG000000870_02500 | |||||||||||
| CAZy Family | CBM47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17363; End: 21721 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM47 | 489 | 599 | 8.5e-16 | 0.8984375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 4.42e-08 | 489 | 614 | 2 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam07691 | PA14 | 2.34e-06 | 753 | 861 | 49 | 138 | PA14 domain. This domain forms an insert in bacterial beta-glucosidases and is found in other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins, including anthrax protective antigen (PA). The domain also occurs in a Dictyostelium prespore-cell-inducing factor Psi and in fibrocystin, the mammalian protein whose mutation leads to polycystic kidney and hepatic disease. The crystal structure of PA shows that this domain (named PA14 after its location in the PA20 pro-peptide) has a beta-barrel structure. The PA14 domain sequence suggests a binding function, rather than a catalytic role. The PA14 domain distribution is compatible with carbohydrate binding. |
| pfam07691 | PA14 | 6.98e-06 | 139 | 261 | 31 | 141 | PA14 domain. This domain forms an insert in bacterial beta-glucosidases and is found in other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins, including anthrax protective antigen (PA). The domain also occurs in a Dictyostelium prespore-cell-inducing factor Psi and in fibrocystin, the mammalian protein whose mutation leads to polycystic kidney and hepatic disease. The crystal structure of PA shows that this domain (named PA14 after its location in the PA20 pro-peptide) has a beta-barrel structure. The PA14 domain sequence suggests a binding function, rather than a catalytic role. The PA14 domain distribution is compatible with carbohydrate binding. |
| smart00758 | PA14 | 9.96e-06 | 756 | 861 | 50 | 134 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
| smart00758 | PA14 | 5.78e-05 | 139 | 184 | 29 | 74 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQL43724.1 | 1.73e-134 | 332 | 1452 | 17 | 1134 |
| QHT65774.1 | 4.59e-43 | 111 | 868 | 1242 | 1889 |
| QJX49093.1 | 9.20e-23 | 744 | 868 | 791 | 908 |
| QNF32356.1 | 1.55e-21 | 726 | 868 | 915 | 1056 |
| AXY74080.1 | 1.00e-16 | 332 | 475 | 810 | 953 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4GWI_A | 3.09e-09 | 480 | 575 | 11 | 109 | His62 mutant of the lectin binding domain of lectinolysin complexed with Lewis y [Streptococcus mitis],4GWJ_A His 62 mutant of the lectin binding domain of Lectinolysin complexed with Lewis b [Streptococcus mitis] |
| 3LE0_A | 4.20e-09 | 480 | 575 | 11 | 109 | ChainA, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEG_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEI_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEK_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q766D5 | 9.04e-06 | 321 | 481 | 109 | 270 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase 1 OS=Mus musculus OX=10090 GN=B4galnt4 PE=2 SV=1 |
| Q76KP1 | 9.05e-06 | 321 | 481 | 108 | 269 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase 1 OS=Homo sapiens OX=9606 GN=B4GALNT4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
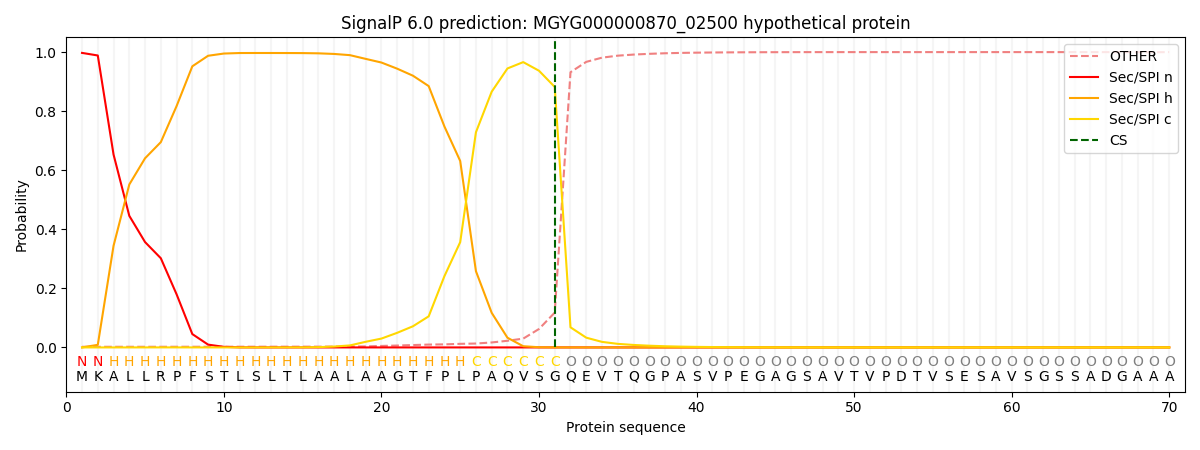
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004140 | 0.994209 | 0.000938 | 0.000267 | 0.000218 | 0.000202 |
