You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000872_01623
You are here: Home > Sequence: MGYG000000872_01623
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA7173 sp900540205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; UBA7173 sp900540205 | |||||||||||
| CAZyme ID | MGYG000000872_01623 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 59161; End: 61911 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.19e-20 | 85 | 420 | 85 | 413 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 4.47e-13 | 189 | 290 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 8.39e-07 | 79 | 310 | 63 | 296 | beta-D-glucuronidase; Provisional |
| PRK09525 | lacZ | 1.25e-06 | 165 | 311 | 196 | 355 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVJ82312.1 | 7.82e-315 | 22 | 915 | 7 | 877 |
| QRP59794.1 | 6.52e-302 | 9 | 915 | 4 | 929 |
| QQT79911.1 | 6.52e-302 | 9 | 915 | 4 | 929 |
| ASM65962.1 | 7.73e-302 | 9 | 915 | 9 | 934 |
| QUU08147.1 | 5.26e-301 | 9 | 915 | 4 | 929 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D8K_A | 3.34e-07 | 79 | 374 | 96 | 374 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 6NCY_A | 3.38e-07 | 128 | 413 | 150 | 416 | Crystalstructure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans [Fusicatenibacter saccharivorans],6NCY_B Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans [Fusicatenibacter saccharivorans],6NCZ_A Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans],6NCZ_B Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans],6NCZ_C Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans],6NCZ_D Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans],6NCZ_E Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans],6NCZ_F Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans bound to phenyl-thio-beta-D-glucuronide [Fusicatenibacter saccharivorans] |
| 7VQM_A | 1.74e-06 | 130 | 413 | 141 | 409 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 6D1P_A | 7.72e-06 | 249 | 413 | 261 | 428 | Apostructure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii],6D1P_B Apo structure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2XQU3 | 2.22e-08 | 165 | 491 | 197 | 545 | Beta-galactosidase 2 OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| Q47077 | 5.02e-08 | 165 | 491 | 197 | 545 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| A4W7D2 | 1.14e-07 | 253 | 416 | 292 | 466 | Beta-galactosidase OS=Enterobacter sp. (strain 638) OX=399742 GN=lacZ PE=3 SV=1 |
| A7FH78 | 1.15e-07 | 85 | 491 | 131 | 559 | Beta-galactosidase OS=Yersinia pseudotuberculosis serotype O:1b (strain IP 31758) OX=349747 GN=lacZ PE=3 SV=1 |
| Q1C6T8 | 1.50e-07 | 85 | 491 | 131 | 559 | Beta-galactosidase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
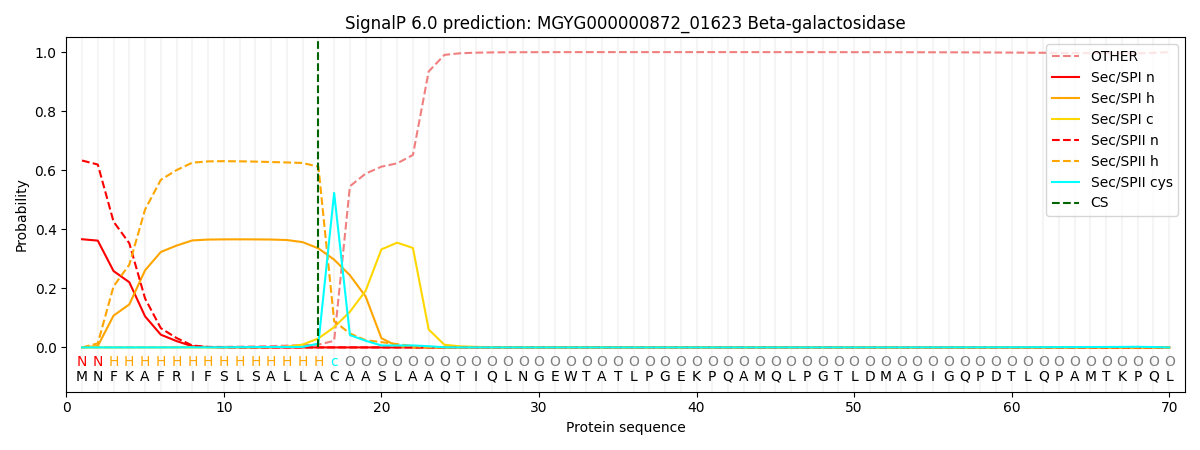
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002347 | 0.353899 | 0.642509 | 0.000510 | 0.000379 | 0.000353 |
