You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000902_00481
Basic Information
help
| Species |
Clostridium neonatale
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium neonatale
|
| CAZyme ID |
MGYG000000902_00481
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000902 |
4770522 |
MAG |
China |
Asia |
|
| Gene Location |
Start: 8213;
End: 10222
Strand: +
|
No EC number prediction in MGYG000000902_00481.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
18 |
242 |
1.3e-39 |
0.40555555555555556 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
2.15e-39 |
26 |
254 |
9 |
234 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
4.03e-30 |
80 |
232 |
1 |
151 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG4745
|
COG4745 |
6.58e-07 |
83 |
202 |
67 |
184 |
Predicted membrane-bound mannosyltransferase [General function prediction only]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O34575
|
2.20e-106 |
25 |
665 |
10 |
677 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
|
P37483
|
3.12e-99 |
27 |
662 |
10 |
661 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000008
|
0.000003
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
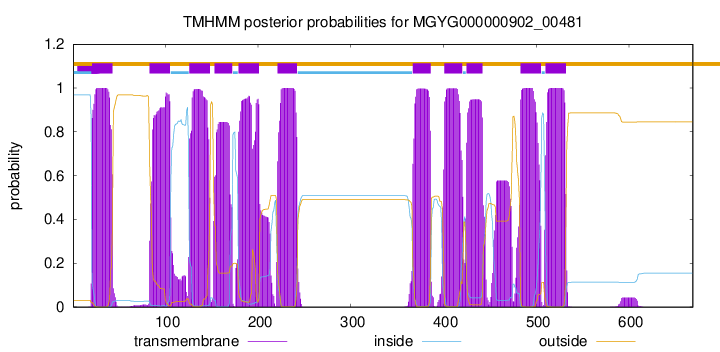
| start |
end |
| 21 |
43 |
| 83 |
105 |
| 126 |
148 |
| 153 |
172 |
| 179 |
201 |
| 221 |
242 |
| 367 |
386 |
| 401 |
420 |
| 425 |
442 |
| 483 |
505 |
| 510 |
532 |


