You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000902_02664
You are here: Home > Sequence: MGYG000000902_02664
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
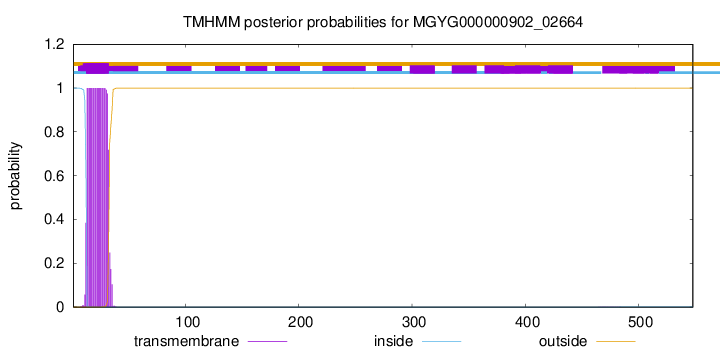
TMHMM annotations
Basic Information help
| Species | Clostridium neonatale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium neonatale | |||||||||||
| CAZyme ID | MGYG000000902_02664 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | Thiol-disulfide oxidoreductase ResA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34497; End: 36143 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 225 | 536 | 2.3e-84 | 0.9906542056074766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02966 | TlpA_like_family | 4.55e-25 | 89 | 201 | 8 | 116 | TlpA-like family; composed of TlpA, ResA, DsbE and similar proteins. TlpA, ResA and DsbE are bacterial protein disulfide reductases with important roles in cytochrome maturation. They are membrane-anchored proteins with a soluble TRX domain containing a CXXC motif located in the periplasm. The TRX domains of this family contain an insert, approximately 25 residues in length, which correspond to an extra alpha helix and a beta strand when compared with TRX. TlpA catalyzes an essential reaction in the biogenesis of cytochrome aa3, while ResA and DsbE are essential proteins in cytochrome c maturation. Also included in this family are proteins containing a TlpA-like TRX domain with domain architectures similar to E. coli DipZ protein, and the N-terminal TRX domain of PilB protein from Neisseria which acts as a disulfide reductase that can recylce methionine sulfoxide reductases. |
| pfam00578 | AhpC-TSA | 3.86e-15 | 86 | 195 | 11 | 121 | AhpC/TSA family. This family contains proteins related to alkyl hydroperoxide reductase (AhpC) and thiol specific antioxidant (TSA). |
| cd03010 | TlpA_like_DsbE | 1.25e-13 | 93 | 203 | 18 | 121 | TlpA-like family, DsbE (also known as CcmG and CycY) subfamily; DsbE is a membrane-anchored, periplasmic TRX-like reductase containing a CXXC motif that specifically donates reducing equivalents to apocytochrome c via CcmH, another cytochrome c maturation (Ccm) factor with a redox active CXXC motif. Assembly of cytochrome c requires the ligation of heme to reduced thiols of the apocytochrome. In bacteria, this assembly occurs in the periplasm. The reductase activity of DsbE in the oxidizing environment of the periplasm is crucial in the maturation of cytochrome c. |
| pfam13905 | Thioredoxin_8 | 1.47e-13 | 101 | 195 | 1 | 94 | Thioredoxin-like. Thioredoxins are small enzymes that participate in redox reactions, via the reversible oxidation of an active centre disulfide bond. |
| PRK03147 | PRK03147 | 6.74e-13 | 78 | 195 | 39 | 151 | thiol-disulfide oxidoreductase ResA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSX01740.1 | 4.21e-153 | 76 | 541 | 56 | 521 |
| QUF82251.1 | 4.21e-153 | 76 | 541 | 56 | 521 |
| QJU43754.1 | 4.21e-153 | 76 | 541 | 56 | 521 |
| AXB84457.1 | 4.96e-153 | 76 | 541 | 61 | 526 |
| QCJ05190.1 | 4.96e-153 | 76 | 541 | 61 | 526 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 1.98e-39 | 247 | 541 | 55 | 350 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 4NMU_A | 7.37e-15 | 82 | 202 | 17 | 132 | CrystalStructure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_B Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_C Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_D Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis] |
| 3ERW_A | 3.48e-10 | 89 | 201 | 24 | 133 | CrystalStructure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_B Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_C Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_D Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_E Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_F Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis],3ERW_G Crystal Structure of StoA from Bacillus subtilis [Bacillus subtilis] |
| 3C73_A | 5.78e-10 | 82 | 201 | 5 | 119 | ChainA, Thiol-disulfide oxidoreductase resA [unidentified],3C73_B Chain B, Thiol-disulfide oxidoreductase resA [unidentified] |
| 3KCM_A | 1.06e-09 | 89 | 201 | 17 | 125 | Thecrystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15],3KCM_B The crystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15],3KCM_C The crystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15],3KCM_D The crystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15],3KCM_E The crystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15],3KCM_F The crystal structure of thioredoxin protein from Geobacter metallireducens [Geobacter metallireducens GS-15] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 2.63e-22 | 225 | 539 | 45 | 355 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
| Q81FU5 | 2.16e-14 | 82 | 202 | 43 | 158 | Thiol-disulfide oxidoreductase ResA OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=resA PE=3 SV=1 |
| A0RBT0 | 2.93e-14 | 82 | 202 | 43 | 158 | Thiol-disulfide oxidoreductase ResA OS=Bacillus thuringiensis (strain Al Hakam) OX=412694 GN=resA PE=3 SV=1 |
| Q81SZ9 | 2.93e-14 | 82 | 202 | 43 | 158 | Thiol-disulfide oxidoreductase ResA OS=Bacillus anthracis OX=1392 GN=resA PE=1 SV=1 |
| Q73B22 | 3.43e-13 | 82 | 215 | 43 | 167 | Thiol-disulfide oxidoreductase ResA OS=Bacillus cereus (strain ATCC 10987 / NRS 248) OX=222523 GN=resA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.976624 | 0.008533 | 0.013840 | 0.000074 | 0.000038 | 0.000921 |