You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000902_04023
You are here: Home > Sequence: MGYG000000902_04023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
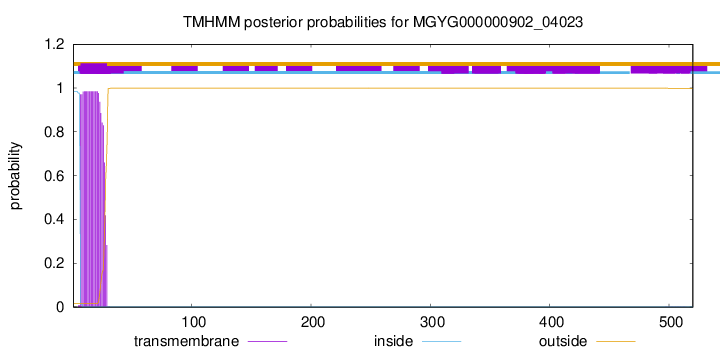
TMHMM annotations
Basic Information help
| Species | Clostridium neonatale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium neonatale | |||||||||||
| CAZyme ID | MGYG000000902_04023 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 235; End: 1797 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 197 | 510 | 5.9e-85 | 0.9875389408099688 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02966 | TlpA_like_family | 3.75e-06 | 63 | 173 | 1 | 116 | TlpA-like family; composed of TlpA, ResA, DsbE and similar proteins. TlpA, ResA and DsbE are bacterial protein disulfide reductases with important roles in cytochrome maturation. They are membrane-anchored proteins with a soluble TRX domain containing a CXXC motif located in the periplasm. The TRX domains of this family contain an insert, approximately 25 residues in length, which correspond to an extra alpha helix and a beta strand when compared with TRX. TlpA catalyzes an essential reaction in the biogenesis of cytochrome aa3, while ResA and DsbE are essential proteins in cytochrome c maturation. Also included in this family are proteins containing a TlpA-like TRX domain with domain architectures similar to E. coli DipZ protein, and the N-terminal TRX domain of PilB protein from Neisseria which acts as a disulfide reductase that can recylce methionine sulfoxide reductases. |
| pfam00578 | AhpC-TSA | 1.12e-05 | 63 | 171 | 7 | 124 | AhpC/TSA family. This family contains proteins related to alkyl hydroperoxide reductase (AhpC) and thiol specific antioxidant (TSA). |
| cd02947 | TRX_family | 0.005 | 104 | 185 | 30 | 92 | TRX family; composed of two groups: Group I, which includes proteins that exclusively encode a TRX domain; and Group II, which are composed of fusion proteins of TRX and additional domains. Group I TRX is a small ancient protein that alter the redox state of target proteins via the reversible oxidation of an active site dithiol, present in a CXXC motif, partially exposed at the protein's surface. TRX reduces protein disulfide bonds, resulting in a disulfide bond at its active site. Oxidized TRX is converted to the active form by TRX reductase, using reducing equivalents derived from either NADPH or ferredoxins. By altering their redox state, TRX regulates the functions of at least 30 target proteins, some of which are enzymes and transcription factors. It also plays an important role in the defense against oxidative stress by directly reducing hydrogen peroxide and certain radicals, and by serving as a reductant for peroxiredoxins. At least two major types of functional TRXs have been reported in most organisms; in eukaryotes, they are located in the cytoplasm and the mitochondria. Higher plants contain more types (at least 20 TRX genes have been detected in the genome of Arabidopsis thaliana), two of which (types f amd m) are located in the same compartment, the chloroplast. Also included in the alignment are TRX-like domains which show sequence homology to TRX but do not contain the redox active CXXC motif. Group II proteins, in addition to either a redox active TRX or a TRX-like domain, also contain additional domains, which may or may not possess homology to known proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APF21752.1 | 7.88e-162 | 1 | 516 | 1 | 521 |
| QSX01740.1 | 4.49e-161 | 1 | 516 | 1 | 521 |
| QUF82251.1 | 4.49e-161 | 1 | 516 | 1 | 521 |
| QJU43754.1 | 4.49e-161 | 1 | 516 | 1 | 521 |
| AXB84457.1 | 5.30e-161 | 1 | 516 | 6 | 526 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 3.81e-43 | 183 | 511 | 13 | 345 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 6R2M_A | 6.61e-16 | 212 | 482 | 33 | 295 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 5.57e-22 | 197 | 509 | 45 | 350 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
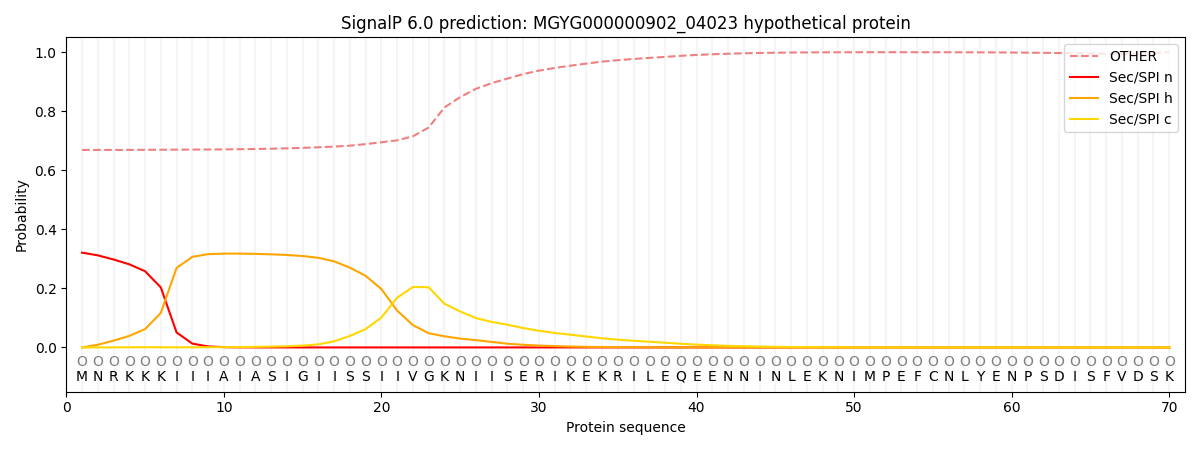
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.684306 | 0.301144 | 0.010768 | 0.001159 | 0.000598 | 0.002026 |