You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000915_02211
You are here: Home > Sequence: MGYG000000915_02211
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium nigeriense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium nigeriense | |||||||||||
| CAZyme ID | MGYG000000915_02211 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11914; End: 14895 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033435 | S-layer_Clost | 8.20e-29 | 580 | 980 | 261 | 722 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
| pfam04122 | CW_binding_2 | 3.63e-20 | 792 | 880 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 6.02e-18 | 694 | 777 | 3 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| COG4193 | LytD | 7.23e-16 | 413 | 580 | 59 | 234 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG2247 | LytB | 2.07e-14 | 701 | 831 | 17 | 147 | Putative cell wall-binding domain [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSW17879.1 | 4.57e-272 | 250 | 993 | 35 | 770 |
| ASW42466.1 | 1.60e-164 | 139 | 698 | 84 | 679 |
| QAS62016.1 | 8.42e-153 | 170 | 684 | 57 | 575 |
| AYE33874.1 | 8.42e-153 | 170 | 684 | 57 | 575 |
| CEI73319.1 | 1.07e-150 | 180 | 983 | 58 | 809 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J72_A | 5.99e-58 | 676 | 974 | 114 | 419 | ChainA, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630],5J72_B Chain B, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630] |
| 5J6Q_A | 4.15e-36 | 690 | 985 | 286 | 587 | ChainA, Cell wall binding protein cwp8 [Clostridioides difficile 630] |
| 7ACX_B | 3.26e-31 | 681 | 989 | 32 | 371 | ChainB, S-layer protein [Clostridioides difficile],7ACX_D Chain D, S-layer protein [Clostridioides difficile] |
| 7ACY_B | 4.40e-31 | 681 | 989 | 32 | 371 | ChainB, S-layer protein [Clostridioides difficile 630],7ACY_D Chain D, S-layer protein [Clostridioides difficile 630],7QGQ_B Chain B, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_D Chain D, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_J Chain J, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_K Chain K, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_L Chain L, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_M Chain M, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_N Chain N, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_T Chain T, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_U Chain U, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_V Chain V, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_W Chain W, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_X Chain X, Precursor of the S-layer proteins [Clostridioides difficile 630] |
| 7ACZ_B | 1.91e-28 | 681 | 989 | 32 | 412 | ChainB, SLPH (HMW SLP) [Clostridioides difficile R20291],7ACZ_D Chain D, SLPH (HMW SLP) [Clostridioides difficile R20291] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02114 | 1.31e-33 | 695 | 951 | 33 | 276 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| P59205 | 3.76e-24 | 433 | 580 | 515 | 656 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 4.38e-24 | 433 | 580 | 559 | 700 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| Q02113 | 1.64e-22 | 695 | 948 | 67 | 302 | Amidase enhancer OS=Bacillus subtilis (strain 168) OX=224308 GN=lytB PE=1 SV=1 |
| O58348 | 1.14e-07 | 714 | 872 | 29 | 186 | Uncharacterized protein PH0614 OS=Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3) OX=70601 GN=PH0614 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
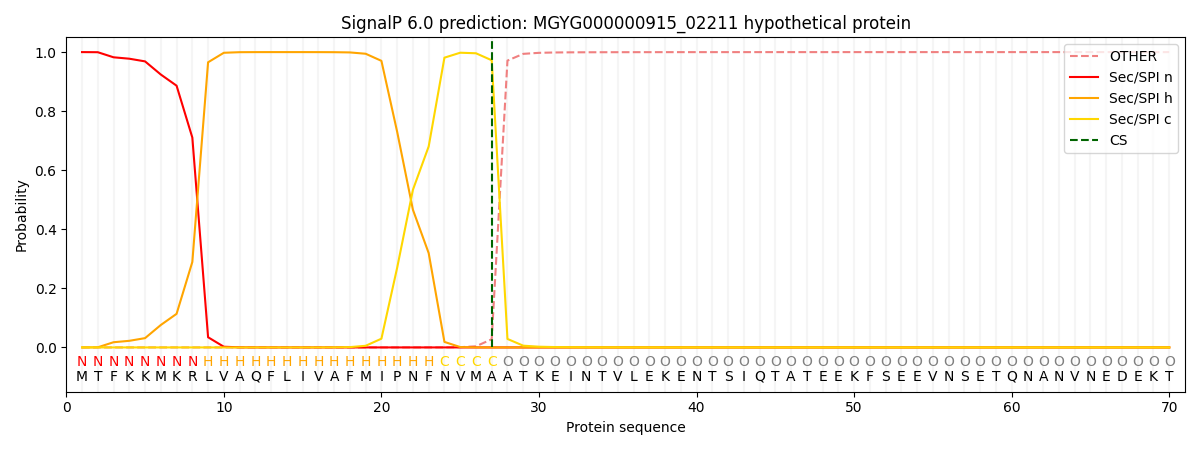
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000388 | 0.998862 | 0.000264 | 0.000157 | 0.000162 | 0.000143 |
