You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000916_01426
You are here: Home > Sequence: MGYG000000916_01426
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactobacillus porci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus porci | |||||||||||
| CAZyme ID | MGYG000000916_01426 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10816; End: 12660 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 233 | 410 | 9.2e-31 | 0.9705882352941176 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06423 | CESA_like | 6.72e-53 | 234 | 412 | 1 | 168 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| cd06434 | GT2_HAS | 1.94e-46 | 231 | 474 | 1 | 231 | Hyaluronan synthases catalyze polymerization of hyaluronan. Hyaluronan synthases (HASs) are bi-functional glycosyltransferases that catalyze polymerization of hyaluronan. HASs transfer both GlcUA and GlcNAc in beta-(1,3) and beta-(1,4) linkages, respectively to the hyaluronan chain using UDP-GlcNAc and UDP-GlcUA as substrates. HA is made as a free glycan, not attached to a protein or lipid. HASs do not need a primer for HA synthesis; they initiate HA biosynthesis de novo with only UDP-GlcNAc, UDP-GlcUA, and Mg2+. Hyaluronan (HA) is a linear heteropolysaccharide composed of (1-3)-linked beta-D-GlcUA-beta-D-GlcNAc disaccharide repeats. It can be found in vertebrates and a few microbes and is typically on the cell surface or in the extracellular space, but is also found inside mammalian cells. Hyaluronan has several physiochemical and biological functions such as space filling, lubrication, and providing a hydrated matrix through which cells can migrate. |
| COG1215 | BcsA | 5.28e-41 | 194 | 581 | 18 | 380 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06439 | CESA_like_1 | 5.61e-38 | 213 | 410 | 10 | 193 | CESA_like_1 is a member of the cellulose synthase (CESA) superfamily. This is a subfamily of cellulose synthase (CESA) superfamily. CESA superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members of the superfamily include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. |
| pfam00535 | Glycos_transf_2 | 8.30e-29 | 233 | 410 | 1 | 164 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKN29834.1 | 7.52e-248 | 11 | 612 | 16 | 621 |
| AKA69493.1 | 1.51e-247 | 11 | 609 | 16 | 619 |
| AWI04409.1 | 6.12e-247 | 11 | 609 | 16 | 619 |
| ANF15068.1 | 8.70e-244 | 11 | 598 | 6 | 597 |
| AOR95077.1 | 1.28e-243 | 11 | 598 | 6 | 598 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SP7_A | 4.44e-29 | 304 | 586 | 171 | 458 | ChainA, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2] |
| 7SP6_A | 1.92e-28 | 304 | 586 | 171 | 458 | ChainA, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SP8_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SP9_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2],7SPA_A Chain A, Hyaluronan synthase [Paramecium bursaria Chlorella virus CZ-2] |
| 6YV7_B | 2.26e-10 | 220 | 460 | 30 | 251 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
| 6YV7_A | 2.27e-10 | 220 | 460 | 31 | 252 | MannosyltransferasePcManGT from Pyrobaculum calidifontis [Pyrobaculum calidifontis JCM 11548],6YV8_A Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP and Mn2+ [Pyrobaculum calidifontis JCM 11548],6YV9_B Mannosyltransferase PcManGT from Pyrobaculum calidifontis in complex with GDP-Man and Mn2+ [Pyrobaculum calidifontis JCM 11548] |
| 6P61_A | 7.12e-06 | 230 | 414 | 13 | 184 | Structureof a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_B Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_C Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197],6P61_D Structure of a Glycosyltransferase from Leptospira borgpetersenii serovar Hardjo-bovis (strain JB197) [Leptospira borgpetersenii serovar Hardjo-bovis str. JB197] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P04340 | 4.10e-38 | 220 | 482 | 41 | 291 | N-acetylglucosaminyltransferase OS=Rhizobium leguminosarum bv. viciae OX=387 GN=nodC PE=3 SV=1 |
| P0DB60 | 1.31e-37 | 201 | 480 | 36 | 307 | Hyaluronan synthase OS=Streptococcus pyogenes serotype M3 (strain ATCC BAA-595 / MGAS315) OX=198466 GN=hasA PE=3 SV=1 |
| P0DB61 | 1.31e-37 | 201 | 480 | 36 | 307 | Hyaluronan synthase OS=Streptococcus pyogenes serotype M3 (strain SSI-1) OX=193567 GN=hasA PE=3 SV=1 |
| Q5X9A9 | 1.31e-37 | 201 | 480 | 36 | 307 | Hyaluronan synthase OS=Streptococcus pyogenes serotype M6 (strain ATCC BAA-946 / MGAS10394) OX=286636 GN=hasA PE=3 SV=1 |
| P50356 | 4.33e-37 | 224 | 482 | 43 | 292 | N-acetylglucosaminyltransferase OS=Rhizobium galegae OX=399 GN=nodC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
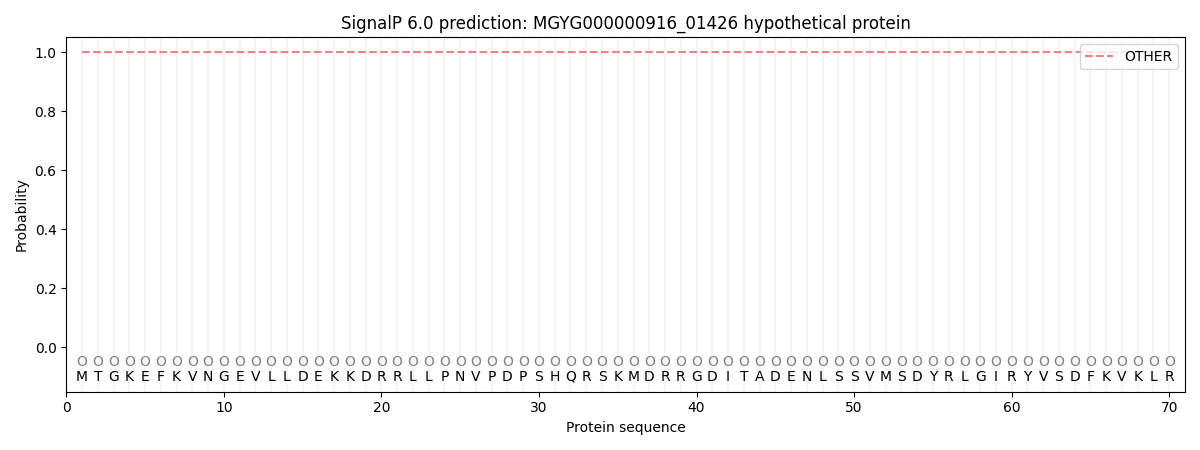
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000055 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
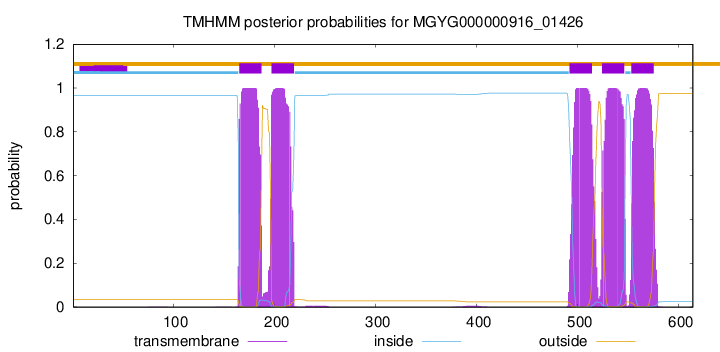
| start | end |
|---|---|
| 165 | 187 |
| 197 | 219 |
| 492 | 514 |
| 524 | 546 |
| 553 | 575 |
