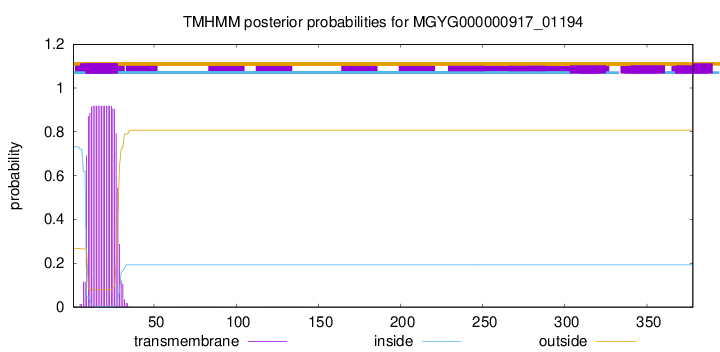You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000917_01194
You are here: Home > Sequence: MGYG000000917_01194
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phascolarctobacterium sp900551745 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Phascolarctobacterium; Phascolarctobacterium sp900551745 | |||||||||||
| CAZyme ID | MGYG000000917_01194 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2313; End: 3449 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 83 | 368 | 7.1e-58 | 0.881619937694704 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBG63434.1 | 1.94e-237 | 1 | 378 | 1 | 378 |
| BBB93524.1 | 2.37e-103 | 11 | 374 | 10 | 371 |
| AFM40559.1 | 1.88e-48 | 81 | 370 | 46 | 334 |
| AVF21138.1 | 2.13e-32 | 83 | 371 | 18 | 305 |
| ARF67512.1 | 2.13e-32 | 83 | 371 | 18 | 305 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 8.17e-17 | 136 | 368 | 111 | 343 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 6R2M_A | 3.95e-13 | 57 | 319 | 25 | 272 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002192 | 0.521626 | 0.474580 | 0.000993 | 0.000350 | 0.000228 |