You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000923_00507
You are here: Home > Sequence: MGYG000000923_00507
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
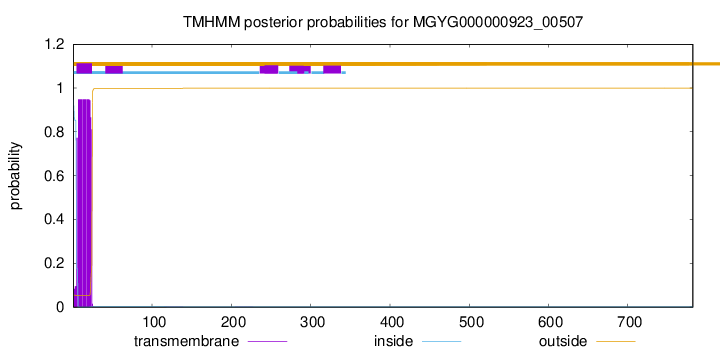
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900552645 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900552645 | |||||||||||
| CAZyme ID | MGYG000000923_00507 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 540350; End: 542698 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 24 | 777 | 5.2e-219 | 0.9899569583931134 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 1.33e-174 | 160 | 504 | 3 | 334 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 2.23e-11 | 668 | 780 | 32 | 159 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF15123.1 | 0.0 | 10 | 780 | 11 | 786 |
| ABR37882.1 | 2.48e-307 | 25 | 780 | 28 | 778 |
| QQY39784.1 | 2.48e-307 | 25 | 780 | 28 | 778 |
| BCA52290.1 | 1.64e-305 | 30 | 781 | 34 | 781 |
| QQY43093.1 | 2.32e-305 | 25 | 780 | 28 | 778 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BY3_A | 3.44e-291 | 30 | 781 | 28 | 775 | Anovel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans [Bacteroides thetaiotaomicron VPI-5482] |
| 7PUG_A | 6.56e-95 | 32 | 619 | 28 | 625 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 7PXQ_A | 1.25e-94 | 32 | 619 | 27 | 624 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 4C90_A | 4.51e-91 | 80 | 619 | 106 | 640 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 4ZMH_A | 9.83e-89 | 108 | 615 | 116 | 606 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
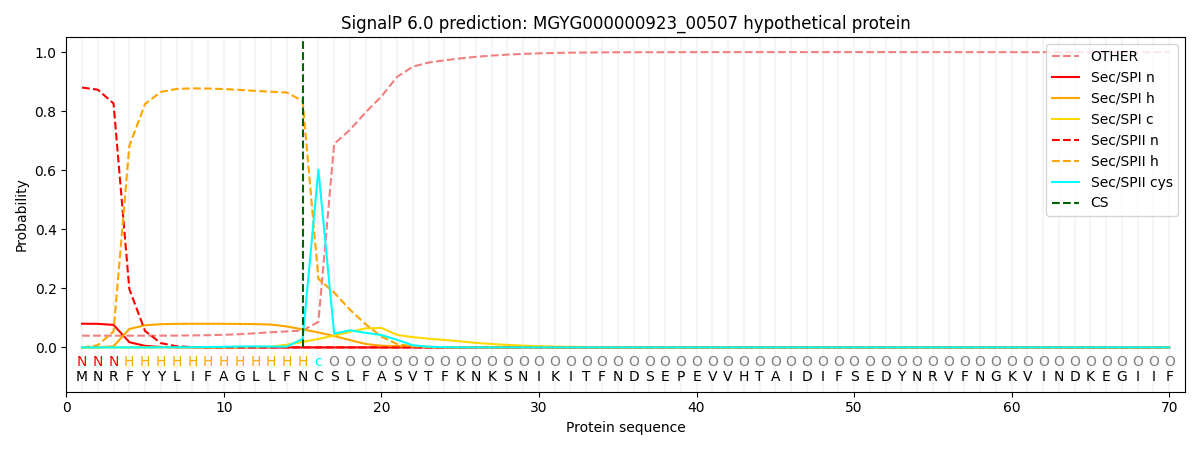
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.040777 | 0.077364 | 0.881578 | 0.000071 | 0.000089 | 0.000125 |