You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000923_02023
You are here: Home > Sequence: MGYG000000923_02023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900552645 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900552645 | |||||||||||
| CAZyme ID | MGYG000000923_02023 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25860; End: 28682 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 18 | 703 | 4.9e-54 | 0.7034574468085106 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.70e-13 | 19 | 433 | 10 | 428 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.49e-09 | 20 | 310 | 11 | 301 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 2.67e-06 | 24 | 155 | 4 | 138 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 0.003 | 200 | 287 | 13 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBE17052.1 | 7.35e-301 | 6 | 940 | 12 | 959 |
| QGY48213.1 | 7.90e-293 | 19 | 940 | 28 | 961 |
| QKH97806.1 | 3.99e-263 | 6 | 940 | 10 | 939 |
| QUT54529.1 | 5.64e-263 | 6 | 940 | 10 | 939 |
| QUT95038.1 | 7.98e-263 | 6 | 940 | 10 | 939 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C70_A | 2.14e-13 | 14 | 418 | 13 | 429 | Thestructure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae],5C70_B The structure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae] |
| 5C71_A | 5.10e-13 | 14 | 418 | 38 | 454 | Thestructure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_B The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_C The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_D The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae] |
| 3GM8_A | 2.13e-11 | 16 | 420 | 1 | 405 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 6D8G_A | 7.76e-10 | 72 | 412 | 73 | 427 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 5UJ6_A | 1.75e-09 | 72 | 412 | 65 | 419 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P08236 | 4.27e-08 | 14 | 515 | 31 | 542 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| Q5R5N6 | 2.21e-07 | 14 | 411 | 31 | 451 | Beta-glucuronidase OS=Pongo abelii OX=9601 GN=GUSB PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
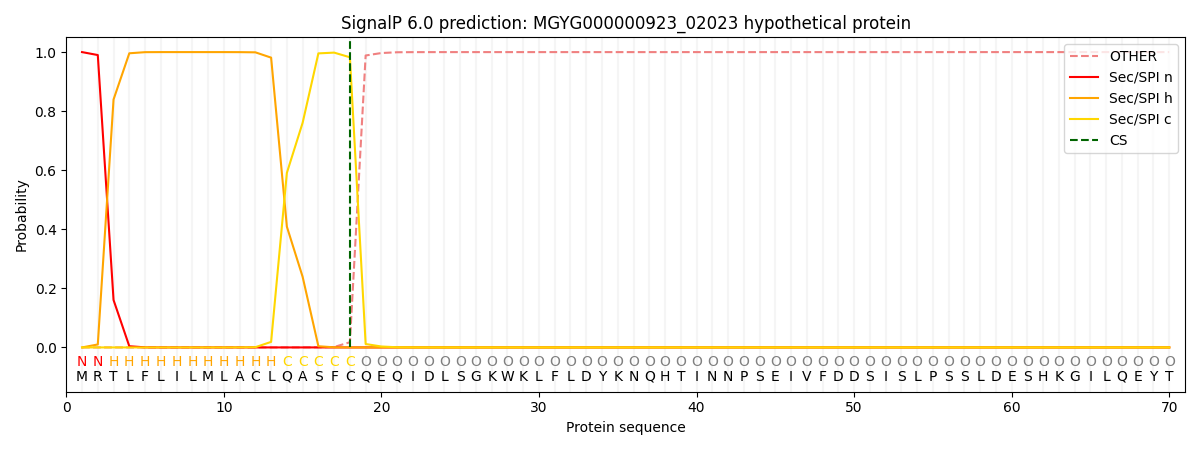
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000186 | 0.999251 | 0.000145 | 0.000155 | 0.000134 | 0.000129 |
