You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000924_00344
You are here: Home > Sequence: MGYG000000924_00344
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | An172 sp002160515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; An172; An172 sp002160515 | |||||||||||
| CAZyme ID | MGYG000000924_00344 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49330; End: 61545 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 2029 | 2183 | 5.9e-20 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 5.79e-25 | 2028 | 2183 | 3 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam00754 | F5_F8_type_C | 9.06e-14 | 2882 | 3008 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| smart00776 | NPCBM | 1.51e-12 | 2023 | 2184 | 1 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 2.03e-12 | 106 | 240 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 6.75e-08 | 1624 | 1741 | 9 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35280.1 | 1.81e-212 | 2019 | 3016 | 40 | 993 |
| AQW23377.1 | 7.16e-211 | 2026 | 3016 | 47 | 993 |
| ATD49073.1 | 8.36e-211 | 2026 | 3016 | 52 | 998 |
| BCL58565.1 | 2.90e-140 | 2024 | 3013 | 77 | 1051 |
| QTR94629.1 | 1.80e-137 | 1978 | 3011 | 72 | 1099 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 1.10e-11 | 2871 | 3010 | 11 | 143 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 7JS4_A | 1.03e-10 | 2041 | 2207 | 622 | 774 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JND_A | 1.91e-09 | 1622 | 1747 | 80 | 208 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 3.34e-09 | 1622 | 1747 | 80 | 208 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JFS_A | 3.45e-09 | 1622 | 1747 | 62 | 190 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 1.48e-09 | 2871 | 3015 | 509 | 650 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q2MGH6 | 2.57e-09 | 2894 | 3001 | 1510 | 1613 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 6.46e-08 | 2894 | 3001 | 1510 | 1613 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
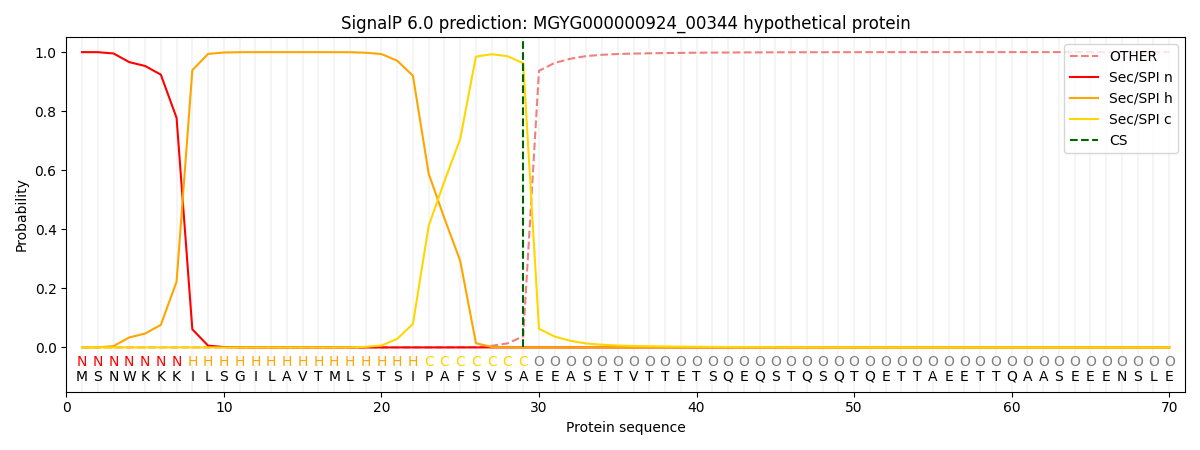
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000526 | 0.998483 | 0.000391 | 0.000217 | 0.000189 | 0.000160 |
