You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000932_01124
You are here: Home > Sequence: MGYG000000932_01124
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS577 sp900543525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS577; UMGS577 sp900543525 | |||||||||||
| CAZyme ID | MGYG000000932_01124 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13643; End: 17572 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH127 | 321 | 543 | 2e-27 | 0.4217557251908397 |
| CBM66 | 902 | 1047 | 6.4e-20 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 2.52e-41 | 107 | 544 | 62 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 3.06e-23 | 282 | 558 | 261 | 508 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| cd14256 | Dockerin_I | 6.66e-07 | 1251 | 1307 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam06439 | DUF1080 | 2.95e-05 | 901 | 1028 | 14 | 143 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPH39723.1 | 9.87e-158 | 44 | 670 | 29 | 649 |
| QJE95537.1 | 6.03e-156 | 44 | 670 | 31 | 648 |
| QNK64903.1 | 9.07e-153 | 21 | 672 | 3 | 647 |
| QHS59113.1 | 1.27e-152 | 41 | 687 | 28 | 667 |
| QIL41453.1 | 6.57e-152 | 44 | 670 | 29 | 649 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
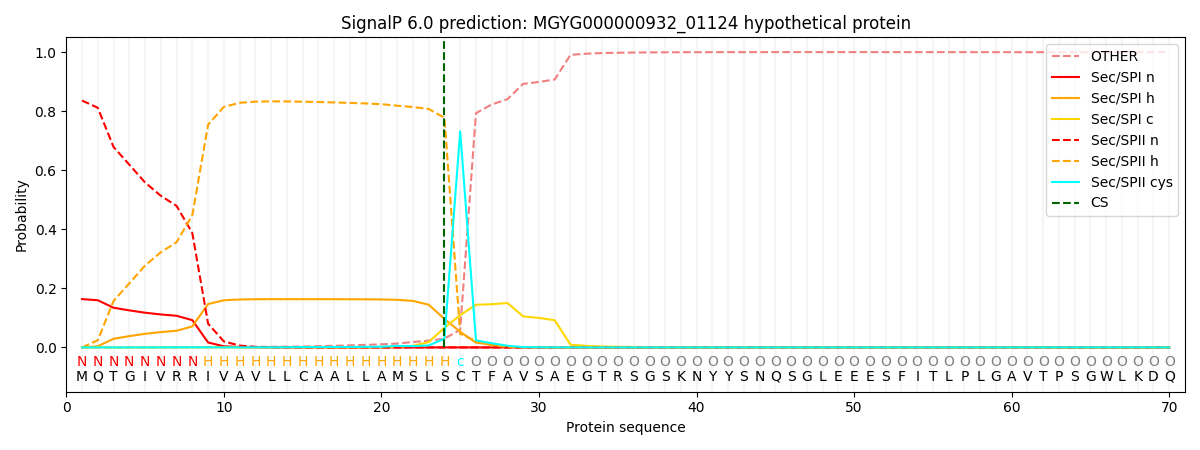
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000483 | 0.161520 | 0.837641 | 0.000216 | 0.000079 | 0.000055 |

