You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000951_02507
You are here: Home > Sequence: MGYG000000951_02507
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066495 sp900545985 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; GCA-900066495; GCA-900066495 sp900545985 | |||||||||||
| CAZyme ID | MGYG000000951_02507 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2713; End: 5601 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.80e-16 | 286 | 466 | 49 | 237 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 7.68e-13 | 344 | 413 | 4 | 73 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| smart00047 | LYZ2 | 4.30e-07 | 335 | 461 | 10 | 143 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG1705 | FlgJ | 1.53e-04 | 345 | 399 | 55 | 106 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| NF038016 | sporang_Gsm | 0.007 | 346 | 383 | 173 | 210 | sporangiospore maturation cell wall hydrolase GsmA. The peptidoglycan-hydrolyzing enzyme GsmA occurs in some sporangia-forming members of the Actinobacteria, such as Actinoplanes missouriensis, and is required for proper separation of spores. GsmA proteins have one or two SH3 domains N-terminal to the hydrolase domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CEI73319.1 | 2.46e-159 | 52 | 546 | 48 | 536 |
| CED94675.1 | 2.85e-152 | 50 | 594 | 49 | 607 |
| QQA11780.1 | 3.95e-123 | 56 | 544 | 73 | 578 |
| AMN34859.1 | 5.71e-123 | 56 | 563 | 73 | 591 |
| ALG48000.1 | 7.25e-123 | 56 | 548 | 73 | 583 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 7.73e-27 | 156 | 452 | 29 | 277 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 3.00e-26 | 156 | 461 | 397 | 654 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 3.58e-26 | 156 | 461 | 441 | 698 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
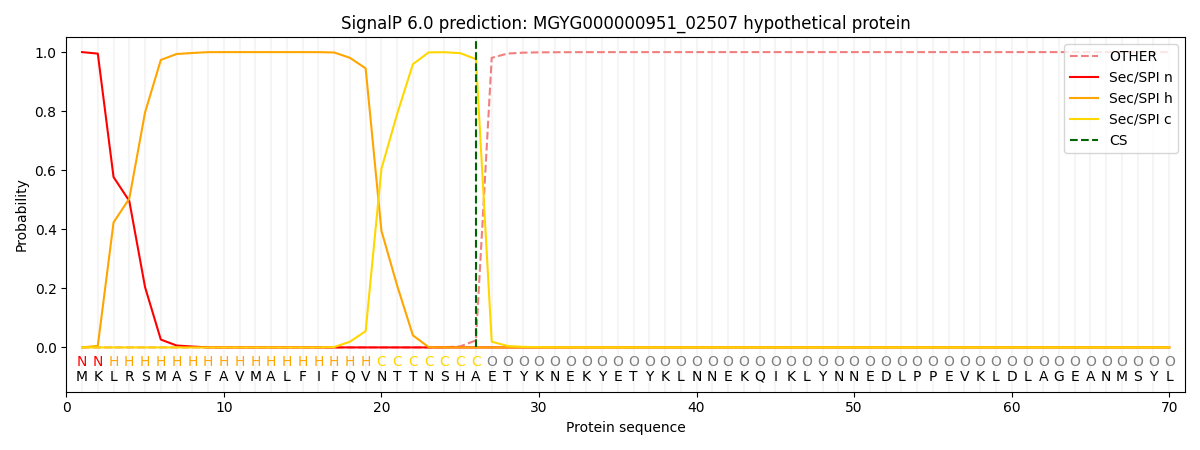
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000283 | 0.999018 | 0.000183 | 0.000175 | 0.000167 | 0.000150 |
