You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000956_00057
You are here: Home > Sequence: MGYG000000956_00057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Barnesiella sp900538705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella sp900538705 | |||||||||||
| CAZyme ID | MGYG000000956_00057 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 60493; End: 62946 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 141 | 335 | 5.2e-40 | 0.8910891089108911 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 4.69e-29 | 180 | 334 | 34 | 186 | Amb_all domain. |
| COG3866 | PelB | 2.09e-28 | 17 | 396 | 20 | 342 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 5.19e-15 | 174 | 334 | 48 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| pfam13205 | Big_5 | 2.20e-04 | 643 | 745 | 9 | 106 | Bacterial Ig-like domain. |
| pfam12733 | Cadherin-like | 0.006 | 430 | 489 | 14 | 85 | Cadherin-like beta sandwich domain. This domain is found in several bacterial, metazoan and chlorophyte algal proteins. A profile-profile comparison recovered the cadherin domain and a comparison of the predicted structure of this domain with the crystal structure of the cadherin showed a congruent seven stranded secondary structure. The domain is widespread in bacteria and seen in the firmicutes, actinobacteria, certain proteobacteria, bacteroides and chlamydiae with an expansion in Clostridium. In contrast, it is limited in its distribution in eukaryotes suggesting that it was derived through lateral transfer from bacteria. In prokaryotes, this domain is widely fused to other domains such as FNIII (Fibronectin Type III), TIG, SLH (S-layer homology), discoidin, cell-wall-binding repeat domain and alpha-amylase-like glycohydrolases. These associations are suggestive of a carbohydrate-binding function for this cadherin-like domain. In animal proteins it is associated with an ATP-grasp domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAW84045.1 | 1.44e-130 | 67 | 404 | 1 | 331 |
| QVJ81552.1 | 3.95e-113 | 22 | 397 | 41 | 421 |
| ADE83376.1 | 7.80e-113 | 22 | 397 | 41 | 421 |
| ABG58437.1 | 2.35e-40 | 108 | 404 | 34 | 343 |
| QOW10430.1 | 5.04e-38 | 131 | 414 | 49 | 352 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VBL_A | 4.04e-15 | 176 | 391 | 146 | 367 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 1PXZ_A | 4.23e-13 | 181 | 336 | 122 | 270 | ChainA, Major pollen allergen Jun a 1 [Juniperus ashei],1PXZ_B Chain B, Major pollen allergen Jun a 1 [Juniperus ashei] |
| 3ZSC_A | 5.45e-12 | 154 | 311 | 63 | 214 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 5AMV_A | 1.18e-07 | 186 | 395 | 150 | 395 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
| 1BN8_A | 1.25e-07 | 186 | 395 | 171 | 416 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5AVN4 | 4.20e-19 | 169 | 398 | 106 | 326 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
| B0XT32 | 1.70e-18 | 187 | 386 | 117 | 311 | Probable pectate lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyA PE=3 SV=1 |
| Q4WIT0 | 1.70e-18 | 187 | 386 | 117 | 311 | Probable pectate lyase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyA PE=3 SV=1 |
| Q9WYR4 | 3.52e-15 | 154 | 311 | 90 | 241 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
| B1L969 | 1.10e-14 | 154 | 311 | 88 | 239 | Pectate trisaccharide-lyase OS=Thermotoga sp. (strain RQ2) OX=126740 GN=pelA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
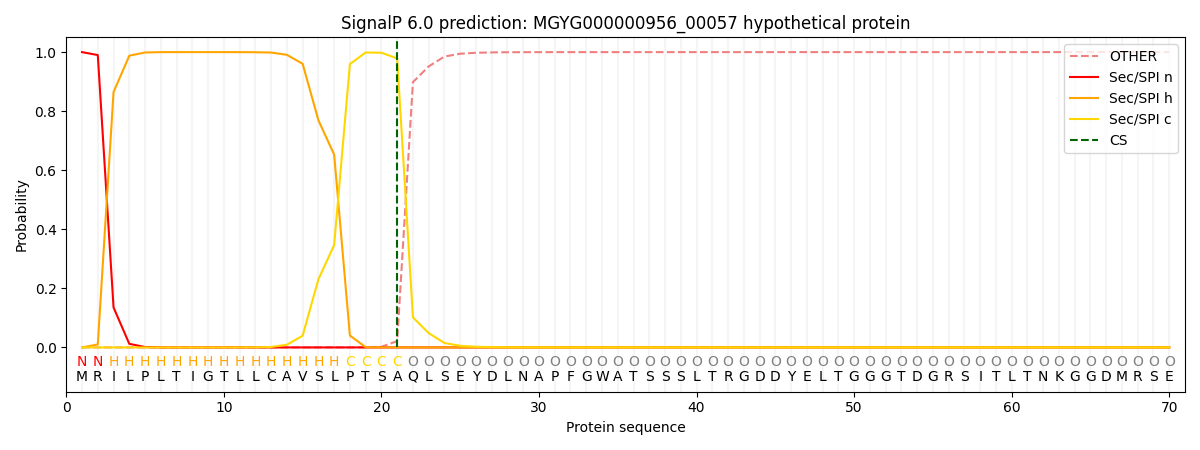
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.998936 | 0.000222 | 0.000184 | 0.000166 | 0.000158 |
