You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000956_01871
You are here: Home > Sequence: MGYG000000956_01871
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Barnesiella sp900538705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella sp900538705 | |||||||||||
| CAZyme ID | MGYG000000956_01871 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | Unsaturated rhamnogalacturonyl hydrolase YteR | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64505; End: 65704 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 46 | 395 | 1.4e-110 | 0.9849397590361446 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 5.30e-104 | 28 | 397 | 1 | 343 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 2.60e-88 | 48 | 396 | 28 | 356 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT73498.1 | 2.03e-182 | 4 | 397 | 8 | 398 |
| CBK68198.1 | 4.49e-182 | 21 | 397 | 9 | 381 |
| SCV08369.1 | 8.23e-182 | 21 | 397 | 26 | 398 |
| QUR41787.1 | 8.23e-182 | 21 | 397 | 26 | 398 |
| QRQ55761.1 | 8.23e-182 | 21 | 397 | 26 | 398 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4WU0_A | 5.18e-97 | 54 | 396 | 23 | 361 | StructuralAnalysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824],4WU0_B Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824] |
| 1NC5_A | 4.52e-71 | 47 | 396 | 32 | 367 | Structureof Protein of Unknown Function of YteR from Bacillus Subtilis [Bacillus subtilis],2D8L_A Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc [Bacillus subtilis] |
| 2GH4_A | 1.89e-70 | 47 | 396 | 22 | 357 | ChainA, Putative glycosyl hydrolase yteR [Bacillus subtilis] |
| 4CE7_A | 1.12e-09 | 159 | 333 | 154 | 323 | Crystalstructure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_B Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_C Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34559 | 2.48e-70 | 47 | 396 | 32 | 367 | Unsaturated rhamnogalacturonyl hydrolase YteR OS=Bacillus subtilis (strain 168) OX=224308 GN=yteR PE=1 SV=1 |
| P0A3U6 | 2.06e-35 | 165 | 392 | 1 | 227 | Protein Atu3128 OS=Agrobacterium fabrum (strain C58 / ATCC 33970) OX=176299 GN=Atu3128 PE=3 SV=1 |
| P0A3U7 | 2.06e-35 | 165 | 392 | 1 | 227 | 24.9 kDa protein in picA locus OS=Rhizobium radiobacter OX=358 PE=2 SV=1 |
| L7P9J4 | 6.31e-09 | 159 | 333 | 161 | 330 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Nonlabens ulvanivorans OX=906888 GN=IL45_01505 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
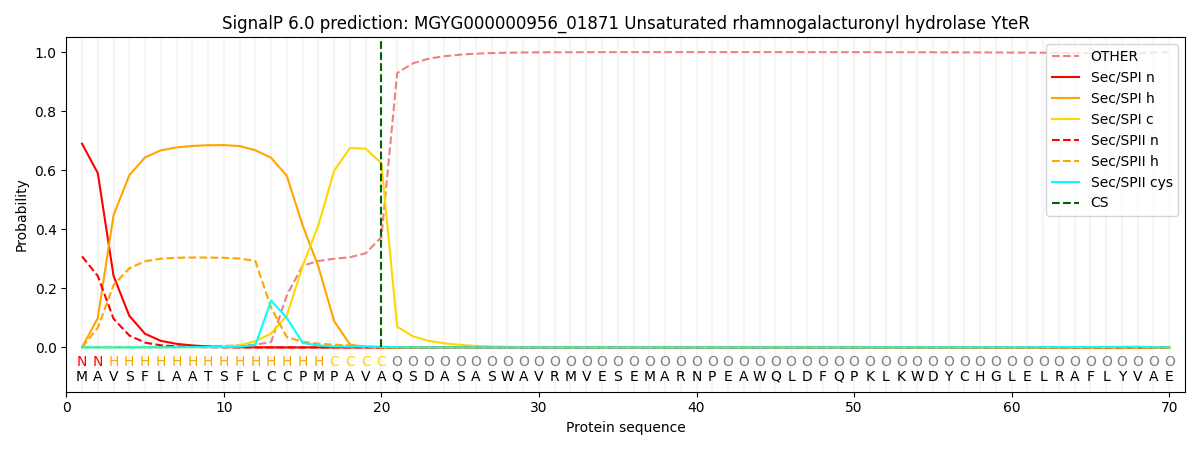
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003280 | 0.676381 | 0.318975 | 0.000616 | 0.000400 | 0.000322 |
